4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
4TYM
 
 | | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935.
To Be Published
|
|
4WCZ
 
 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
3S5S
 
 | |
5DK6
 
 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
4XAP
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 | | Descriptor: | Aldo-keto reductase | | Authors: | Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021
to be published
|
|
4XA8
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding | | Authors: | Handing, K.B, Gasiorowska, O.A, Shabalin, I.G, Cymborowski, M.T, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-12 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2.
to be published
|
|
5C9G
 
 | | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium | | Descriptor: | D-MALATE, Enoyl-CoA hydratase/isomerase family protein, TETRAETHYLENE GLYCOL | | Authors: | Szlachta, K, Cooper, D.R, Chapman, H.C, Cymbrowski, M.T, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, Hammonds, J, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium
to be published
|
|
5C7H
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH | | Descriptor: | Aldo-keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-24 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH
to be published
|
|
4MOU
 
 | | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282 | | Descriptor: | CACODYLATE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Chapman, H.C, Cooper, D.R, Geffken, K.T, Cymborowski, M.T, Osinski, T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282
To be Published
|
|
4MGG
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Muconate lactonizing enzyme, ... | | Authors: | Vetting, M.W, Zhang, X, Wasserman, S.R, Morisco, L.L, Sojitra, S, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121
To be Published
|
|
4OMR
 
 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX in complex with acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, BENZAMIDINE, Enoyl-CoA hydratase | | Authors: | Langner, K.M, Cooper, D.R, Chapman, H.C, Handing, K.B, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-27 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX in complex with acetoacetyl-CoA
To be Published
|
|
4NEK
 
 | | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Geffken, K, Chapman, H.C, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1
To be Published
|
|
4PFQ
 
 | | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 029763. | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 0299763.
to be published
|
|
4RQB
 
 | |
4RQA
 
 | |
4RPF
 
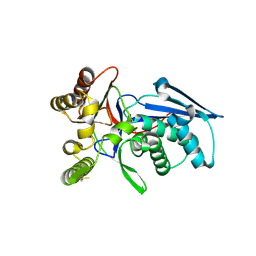 | | Crystal structure of homoserine kinase from Yersinia pestis Nepal516, NYSGRC target 032715 | | Descriptor: | CITRIC ACID, Homoserine kinase | | Authors: | Ptskovsky, Y, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Homoserine Kinase from Yersinia Pestis Nepal516, Nysgrc Target 032715
To be Published
|
|
4DZH
 
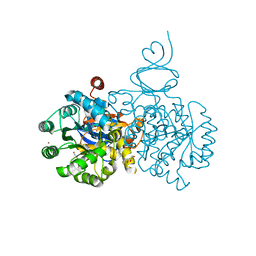 | | Crystal structure of an adenosine deaminase from xanthomonas campestris (target nysgrc-200456) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Crystal structure of an adenosine deaminase from xanthomonas campestris (target nysgrc-200456) with bound zn
to be published
|
|
4DYK
 
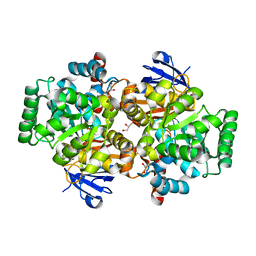 | | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn
to be published
|
|
4G4F
 
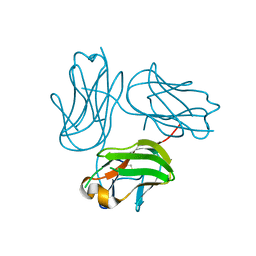 | |
4G8S
 
 | | Crystal Structure Of a Putative Nitroreductase from Geobacter sulfurreducens PCA (Target PSI-013445) | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Nitroreductase family protein | | Authors: | Kumar, P.R, Bhosle, R, Hillerich, B, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nitroreductase from Geobacter sulfurreducens PCA
to be published
|
|
4GQA
 
 | | Crystal structure of NAD binding oxidoreductase from Klebsiella pneumoniae | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAD binding Oxidoreductase, ... | | Authors: | Osinski, S, Majorek, K.A, Niedzialkowska, E, Osinski, T, Porebski, P.J, Nawar, A, Hammonds, J, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-22 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of NAD binding oxidoreductase from Klebsiella pneumoniae (CASP Target)
To be Published
|
|
3RQ3
 
 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form | | Descriptor: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Ramagopal, U.A, Rubinstein, R, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-04-27 | | Release date: | 2011-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form
To be published
|
|
4KA0
 
 | | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P21221 | | Descriptor: | CHLORIDE ION, Putative thiol-disulfide oxidoreductase | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R.D, Bonanno, J.B, Armstrong, R.N, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-21 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P21221
To be Published
|
|
4J2H
 
 | | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708) | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Sampathkumar, P, Gizzi, A, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Stead, M, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708)
to be published
|
|
