4WD8
 
 | |
3KLY
 
 | |
3KLZ
 
 | |
4WD7
 
 | |
6WTW
 
 | | Structure of LaINDY crystallized in the presence of alpha-ketoglutarate and malate | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Cocco, N, Marden, J.J, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WM5
 
 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 3,3',3''-phosphanetriyltripropanoic acid, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-04-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
6WU2
 
 | | Structure of the LaINDY-malate complex | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU4
 
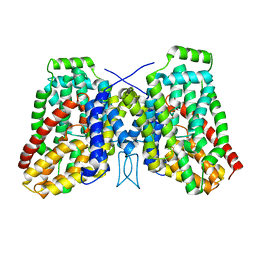 | | Structure of the LaINDY-alpha-ketoglutarate complex | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU1
 
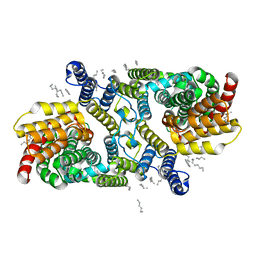 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
5IWS
 
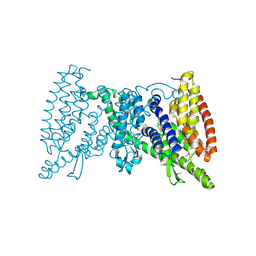 | | Crystal structure of the transporter MalT, the EIIC domain from the maltose-specific phosphotransferase system | | Descriptor: | Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | McCoy, J.G, Ren, Z, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The Structure of a Sugar Transporter of the Glucose EIIC Superfamily Provides Insight into the Elevator Mechanism of Membrane Transport.
Structure, 24, 2016
|
|
5WUF
 
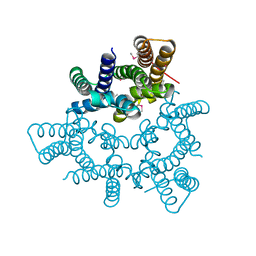 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Mao, Y, Gao, F, Su, M, Wang, X.H, Zeng, Y, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
3LLQ
 
 | |
5WUC
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUD
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUE
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
3M78
 
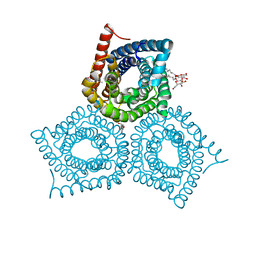 | |
3M74
 
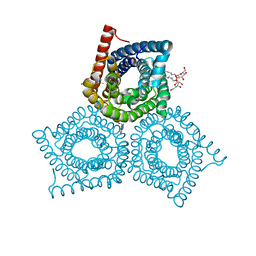 | |
3M7E
 
 | |
3M7L
 
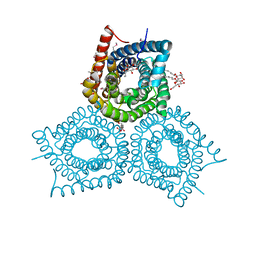 | |
3M77
 
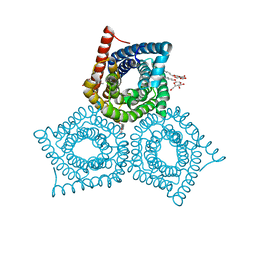 | |
3M7C
 
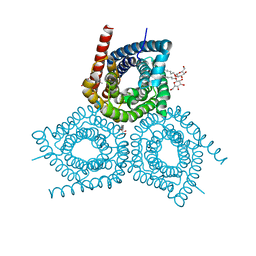 | |
3M71
 
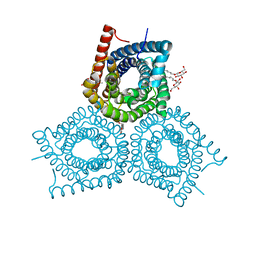 | | Crystal Structure of Plant SLAC1 homolog TehA | | Descriptor: | Tellurite resistance protein tehA homolog, octyl beta-D-glucopyranoside | | Authors: | Chen, Y.-H, Hu, L, Punta, M, Bruni, R, Hillerich, B, Kloss, B, Rost, B, Love, J, Siegelbaum, S.A, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-03-16 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Homologue structure of the SLAC1 anion channel for closing stomata in leaves.
Nature, 467, 2010
|
|
3ME1
 
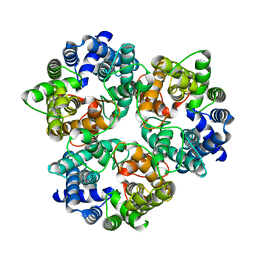 | |
3M75
 
 | |
3M76
 
 | |
