6YSF
 
 | |
6YSL
 
 | |
8FY4
 
 | | Structure of NOT1:NOT10:NOT11 module of the chicken CCR4-NOT complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Lea, S.M, Deme, J.C, Raisch, T, Levdansky, Y, Valkov, E. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structure and assembly of the NOT10:11 module of the CCR4-NOT complex.
Commun Biol, 6, 2023
|
|
8FY3
 
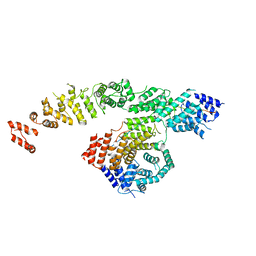 | | Structure of NOT1:NOT10:NOT11 module of the human CCR4-NOT complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Lea, S.M, Deme, J.C, Raisch, T, Pekovic, F, Valkov, E. | | Deposit date: | 2023-01-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure and assembly of the NOT10:11 module of the CCR4-NOT complex.
Commun Biol, 6, 2023
|
|
7B29
 
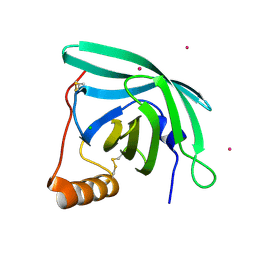 | |
7B2A
 
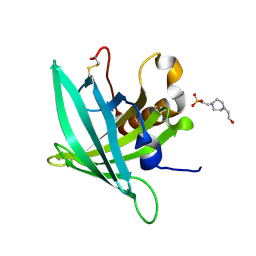 | |
7B2D
 
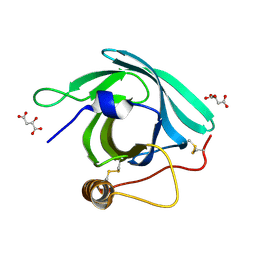 | |
7B26
 
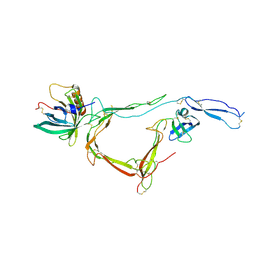 | | CirpA1 in complex with pseudo-monomeric Properdin lacking TSR2-3 | | Descriptor: | CirpA1, Properdin, alpha-D-mannopyranose, ... | | Authors: | Lea, S.M, Johnson, S, Braunger, K. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of a family of tick-derived complement inhibitors targeting properdin.
Nat Commun, 13, 2022
|
|
7B28
 
 | |
6SD6
 
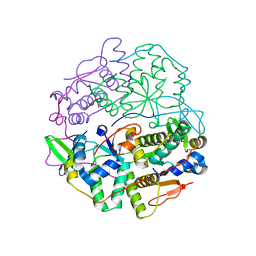 | | Structure of VapBC from Shigella sonnei | | Descriptor: | Antitoxin, tRNA(fMet)-specific endonuclease VapC | | Authors: | Lea, S.M, Hollingshead, S. | | Deposit date: | 2019-07-26 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Polymorphisms in the VapBC toxin:antitoxin system mediate high frequency plasmid loss in Shigella sonnei
To Be Published
|
|
7NVG
 
 | | Salmonella flagellar basal body refined in C1 map | | Descriptor: | Basal-body rod modification protein FlgD, Flagellar L-ring protein, Flagellar M-ring protein, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7NVH
 
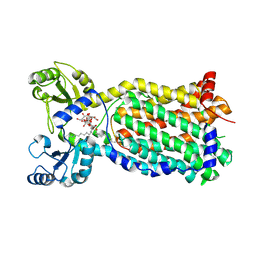 | | Cryo-EM structure of the mycolic acid transporter MmpL3 from M. tuberculosis | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Trehalose monomycolate exporter MmpL3 | | Authors: | Adams, O, Deme, J.C, Parker, J.L, Lea, S.M, Newstead, S. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure and resistance landscape of M. tuberculosis MmpL3: An emergent therapeutic target.
Structure, 29, 2021
|
|
6F2D
 
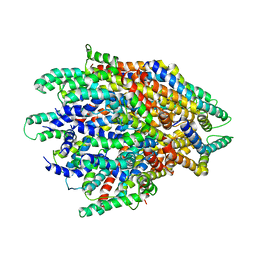 | | A FliPQR complex forms the core of the Salmonella type III secretion system export apparatus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Johnson, S, Kuhlen, L, Abrusci, P, Lea, S.M. | | Deposit date: | 2017-11-24 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the core of the type III secretion system export apparatus.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2XRC
 
 | | Human complement factor I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HUMAN COMPLEMENT FACTOR I | | Authors: | Roversi, P, Johnson, S, Lea, S.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Basis for Complement Factor I Control and its Disease-Associated Sequence Polymorphisms.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1H2Q
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Powell, R, Spiller, O.B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
7SB2
 
 | |
7SAT
 
 | |
7SAZ
 
 | |
7SAU
 
 | |
7SAX
 
 | |
4CUD
 
 | | Human Notch1 EGF domains 11-13 mutant fucosylated at T466 | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, alpha-L-fucopyranose | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CVX
 
 | | COMPLEX OF A B2 CHICKEN MHC CLASS I MOLECULE AND A 9MER CHICKEN PEPTIDE | | Descriptor: | BETA-2-MICROGLOBULIN, MHC CLASS I ALPHA CHAIN 2, SELF-PEPTIDE | | Authors: | Chappell, P.E, Roversi, P, Harrison, M.C, Mears, L.E, Kaufman, J.F, Lea, S.M. | | Deposit date: | 2014-03-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding.
Elife, 4, 2015
|
|
4D0F
 
 | | Human Notch1 EGF domains 11-13 mutant T466A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CUE
 
 | | Human Notch1 EGF domains 11-13 mutant T466V | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CW1
 
 | | COMPLEX OF A B14 CHICKEN MHC CLASS I MOLECULE AND A 9MER CHICKEN PEPTIDE | | Descriptor: | BETA-2-MICROGLOBULIN, MAJOR HISTOCOMPATIBILITY COMPLEX CLASS I GLYCOPROTEIN HAPLOTYPE B14, PEPTIDE | | Authors: | Chappell, P.E, Roversi, P, Harrison, M.C, Mears, L.E, Kaufman, J.F, Lea, S.M. | | Deposit date: | 2014-03-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding.
Elife, 4, 2015
|
|
