1SMA
 
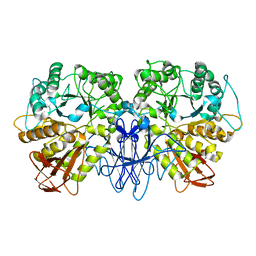 | | CRYSTAL STRUCTURE OF A MALTOGENIC AMYLASE | | Descriptor: | MALTOGENIC AMYLASE | | Authors: | Kim, J.S, Cha, S.S, Oh, B.H. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a maltogenic amylase provides insights into a catalytic versatility.
J.Biol.Chem., 274, 1999
|
|
1T6S
 
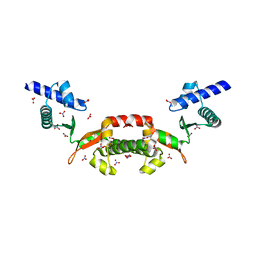 | | Crystal structure of a conserved hypothetical protein from Chlorobium tepidum | | Descriptor: | NITRATE ION, conserved hypothetical protein | | Authors: | Kim, J.S, Shin, D.H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ScpB from Chlorobium tepidum, a protein involved in chromosome partitioning.
Proteins, 62, 2006
|
|
1R6V
 
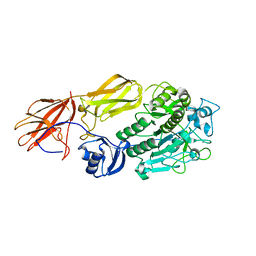 | | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin | | Descriptor: | CALCIUM ION, subtilisin-like serine protease | | Authors: | Kim, J.S, Kluskens, L.D, de Vos, W.M, Huber, R, van der Oost, J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin.
J.Mol.Biol., 335, 2004
|
|
1YF2
 
 | | Three-dimensional structure of DNA sequence specificity (S) subunit of a type I restriction-modification enzyme and its functional implications | | Descriptor: | Type I restriction-modification enzyme, S subunit | | Authors: | Kim, J.S, Degiovanni, A, Jancarik, J, Adams, P.D, Yokota, H.A, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-12-30 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5CHI
 
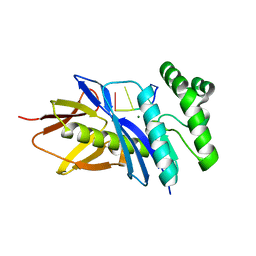 | | Crystal structure of PF2046 in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), MAGNESIUM ION, Uncharacterized protein | | Authors: | Kim, J.S, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural basis of two-nucleotide removal of ssDNA by a cryptic RNase H fold 3'-5' exonuclease PF2046 from Pyrococcus furiosus
to be published
|
|
4WFQ
 
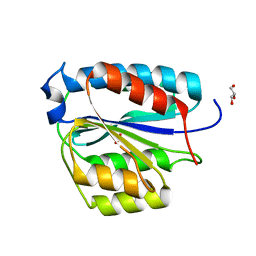 | | Crystal structure of TFIIH subunit | | Descriptor: | GLYCEROL, SULFATE ION, Suppressor of stem-loop protein 1 | | Authors: | Cho, Y, Kim, J.S, Lim, H.S. | | Deposit date: | 2014-09-17 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Rad3/XPD regulatory domain of Ssl1/p44
J.Biol.Chem., 290, 2015
|
|
4QA8
 
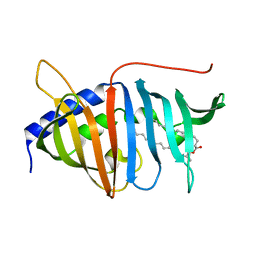 | | Crystal structure of LprF from Mycobacterium bovis | | Descriptor: | (2R)-2-(dodecanoyloxy)propyl (4E,6E,8E,10E,12E)-pentadeca-4,6,8,10,12-pentaenoate, Putative lipoprotein LprF | | Authors: | Ha, N.C, Jiao, L, Kim, J.S. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure and functional implications of LprF from Mycobacterium tuberculosis and M. bovis
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5C0Q
 
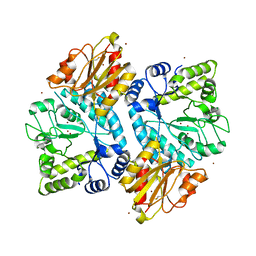 | |
5BZA
 
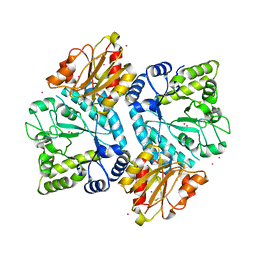 | |
5C22
 
 | | Crystal structure of Zn-bound HlyD from E. coli | | Descriptor: | Chromosomal hemolysin D, ZINC ION | | Authors: | Ha, N.C, Kim, J.S. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
5C21
 
 | | Crystal structure of native HlyD from E. coli | | Descriptor: | Chromosomal hemolysin D | | Authors: | Ha, N.C, Kim, J.S, Yoon, B.Y. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
4Y0M
 
 | |
4XWS
 
 | |
5XSF
 
 | |
8I07
 
 | |
8I08
 
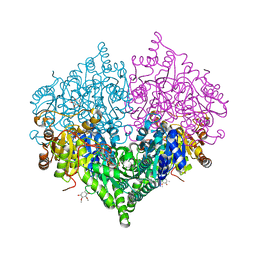 | | Crystal structure of Escherichia coli glyoxylate carboligase quadruple mutant | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8I01
 
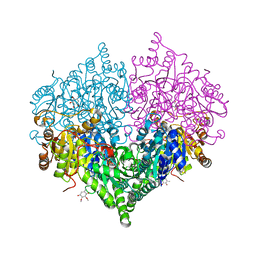 | | Crystal structure of Escherichia coli glyoxylate carboligase | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
8I05
 
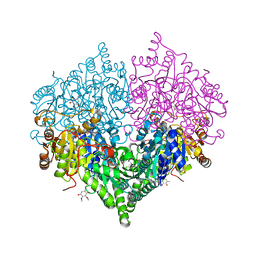 | | Crystal structure of Escherichia coli glyoxylate carboligase double mutant | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Engineering of two thiamine diphosphate-dependent enzymes for the regioselective condensation of C1-formaldehyde into C4-erythrulose.
Int.J.Biol.Macromol., 253, 2023
|
|
7CT6
 
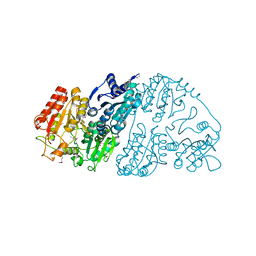 | | Crystal structure of GCL from Deinococcus metallilatus | | Descriptor: | Glyoxylate carboligase | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glyoxylate carboligase-based whole-cell biotransformation of formaldehyde into ethylene glycol via glycolaldehyde.
Green Chem, 1, 2022
|
|
7XZ3
 
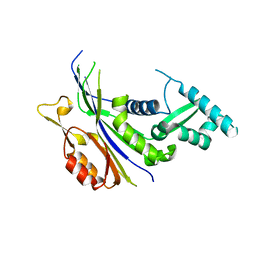 | | Crystal structure of the Type I-B CRISPR-associated protein, Csh2 from Thermobaculum terrenum | | Descriptor: | CRISPR-associated protein, Csh2 family | | Authors: | Seo, P.W, Gu, D.H, Park, S.Y, Kim, J.S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural characterization of the type I-B CRISPR Cas7 from Thermobaculum terrenum.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
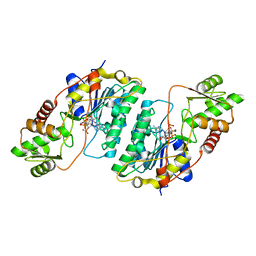
7CXT
 
 | |
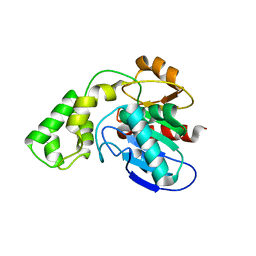
3BF7
 
 | |
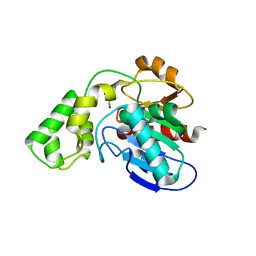
3BF8
 
 | |
3BD5
 
 | |
3BD4
 
 | |
