1C4Z
 
 | | STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY | | Descriptor: | UBIQUITIN CONJUGATING ENZYME E2, UBIQUITIN-PROTEIN LIGASE E3A | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
5NWQ
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with guanidine. | | Descriptor: | GUANIDINE, Guanidine III riboswitch, MAGNESIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-08 | | Release date: | 2017-10-18 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NZD
 
 | | The structure of the thermobifida fusca guanidine III riboswitch in space group P212121. | | Descriptor: | ACETATE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-13 | | Release date: | 2017-10-18 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NZ3
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-12 | | Release date: | 2017-10-18 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NY8
 
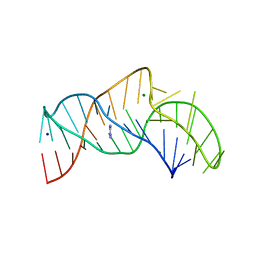 | | The structure of the thermobifida fusca guanidine III riboswitch with aminoguanidine | | Descriptor: | AMINOGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-10-18 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NZ6
 
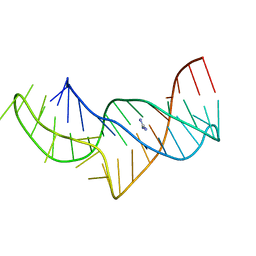 | |
5O62
 
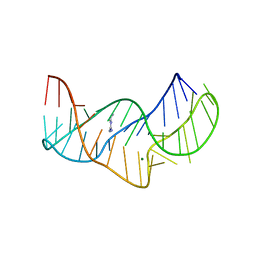 | |
5O69
 
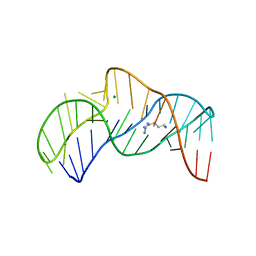 | | The structure of the thermobifida fusca guanidine III riboswitch with agmatine. | | Descriptor: | AGMATINE, MAGNESIUM ION, RNA (37-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-18 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
1LFD
 
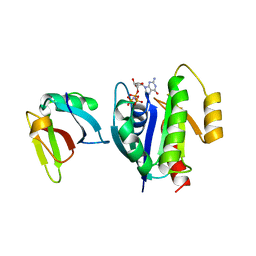 | | CRYSTAL STRUCTURE OF THE ACTIVE RAS PROTEIN COMPLEXED WITH THE RAS-INTERACTING DOMAIN OF RALGDS | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RALGDS, ... | | Authors: | Huang, L, Hofer, F, Martin, G.S, Kim, S.-H. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction of Ras with RalGDS.
Nat.Struct.Biol., 5, 1998
|
|
1YQ3
 
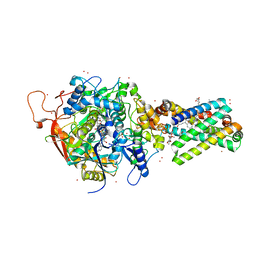 | | Avian respiratory complex ii with oxaloacetate and ubiquinone | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Coenzyme Q10, (2Z,6E,10Z,14E,18E,22E,26Z)-isomer, ... | | Authors: | Huang, L, Cobessi, D, Tung, E.Y, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
1YCY
 
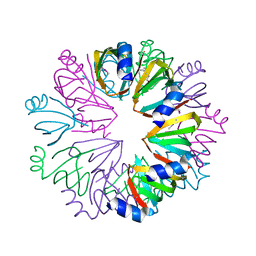 | | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein | | Authors: | Huang, L, Liu, Z.-J, Lee, D, Tempel, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus
To be published
|
|
1LXD
 
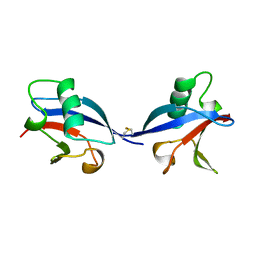 | | CRYSTAL STRUCTURE OF THE RAS INTERACTING DOMAIN OF RALGDS, A GUANINE NUCLEOTIDE DISSOCIATION STIMULATOR OF RAL PROTEIN | | Descriptor: | RALGDSB | | Authors: | Huang, L, Weng, X.W, Hofer, F, Martin, G.S, Kim, S.H. | | Deposit date: | 1997-03-05 | | Release date: | 1998-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the Ras-interacting domain of RalGDS.
Nat.Struct.Biol., 4, 1997
|
|
1YQ4
 
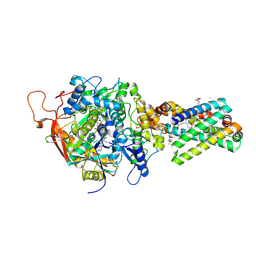 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | Authors: | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
1D5F
 
 | | STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY | | Descriptor: | E6AP HECT CATALYTIC DOMAIN, E3 LIGASE | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-07 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
5LQT
 
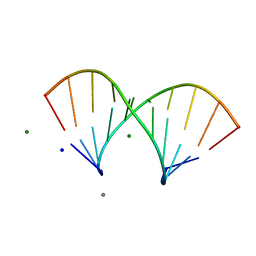 | |
5LR4
 
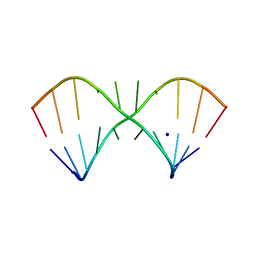 | |
5LQO
 
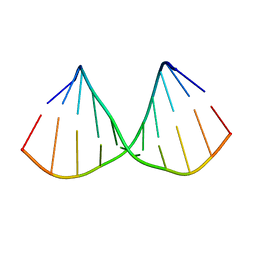 | |
5LR3
 
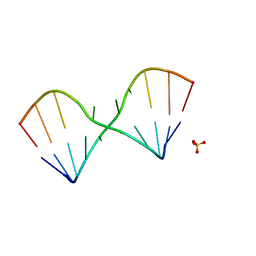 | |
5LR5
 
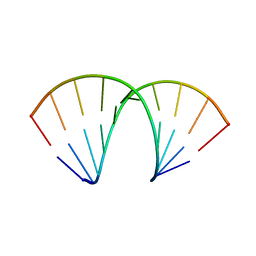 | |
5NEF
 
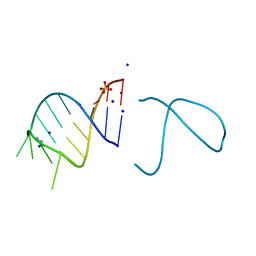 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with guanidine | | Descriptor: | GUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-06-07 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NDH
 
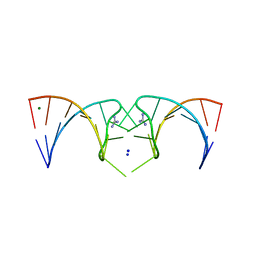 | | The structure of the G. violaceus guanidine II riboswitch P2 stem-loop | | Descriptor: | GUANIDINE, MAGNESIUM ION, RNA (5'-R(*GP*(CBV)P*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*GP*C)-3'), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-31 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NOM
 
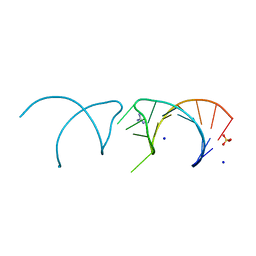 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with guanidine | | Descriptor: | GUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NDI
 
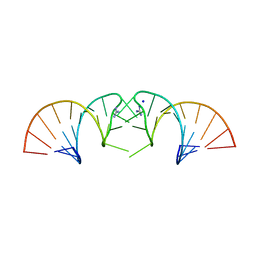 | |
5NEP
 
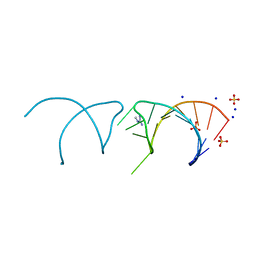 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-31 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NEQ
 
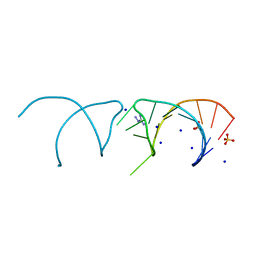 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with aminoguanidine | | Descriptor: | AMINOGUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
