1FTK
 
 | |
1FTM
 
 | |
2NWW
 
 | | Crystal structure of GltPh in complex with TBOA | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, 425aa long hypothetical proton glutamate symport protein | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
4TLL
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 1 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
4TLM
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 2 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
6C13
 
 | | CryoEM structure of mouse PCDH15-4EC-LHFPL5 complex | | Descriptor: | Protocadherin-15, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gouaux, E, Ge, J, Elferich, J. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (11.33 Å) | | Cite: | Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5.
Elife, 7, 2018
|
|
2NWX
 
 | | Crystal structure of GltPh in complex with L-aspartate and sodium ions | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, PALMITIC ACID, ... | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
6C10
 
 | | Crystal structure of mouse PCDH15 EC11-EL | | Descriptor: | Protocadherin-15, alpha-D-mannopyranose | | Authors: | Gouaux, E, Elferich, J, Ge, J. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5.
Elife, 7, 2018
|
|
6C14
 
 | |
2NWL
 
 | | Crystal structure of GltPh in complex with L-Asp | | Descriptor: | ASPARTIC ACID, PALMITIC ACID, glutamate symport protein | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
4XP1
 
 | | X-ray structure of Drosophila dopamine transporter bound to neurotransmitter dopamine | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XP5
 
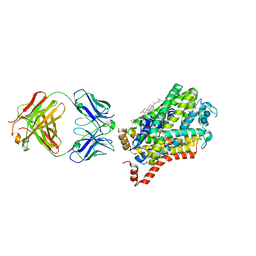 | | X-ray structure of Drosophila dopamine transporter bound to cocaine analogue-RTI55 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Antibody fragment heavy chain-protein, 9D5-heavy chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4M48
 
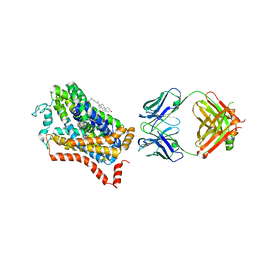 | | X-ray structure of dopamine transporter elucidates antidepressant mechanism | | Descriptor: | 9D5 antibody, heavy chain, light chain, ... | | Authors: | Gouaux, E, Penmatsa, A, Wang, K. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | X-ray structure of dopamine transporter elucidates antidepressant mechanism.
Nature, 503, 2013
|
|
6NJM
 
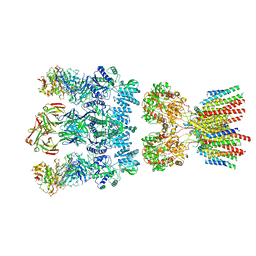 | | Architecture and subunit arrangement of native AMPA receptors | | Descriptor: | 15F1 Fab heavy chain, 15F1 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Zhao, Y. | | Deposit date: | 2019-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM.
Science, 364, 2019
|
|
6NJN
 
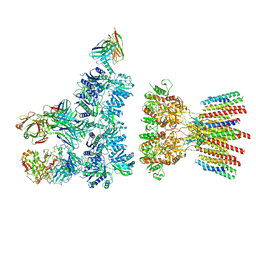 | |
6NJL
 
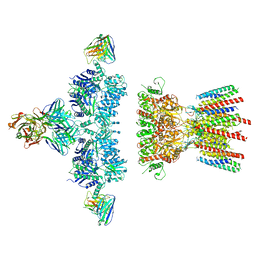 | |
2I3V
 
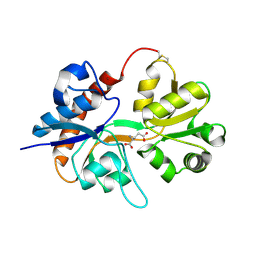 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
8VBY
 
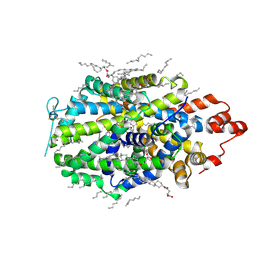 | |
2I3W
 
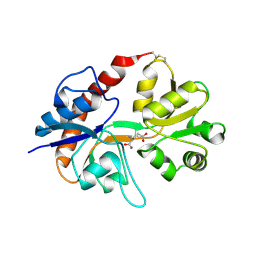 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of S729C mutant | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT 2, GLUTAMIC ACID | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
1LB8
 
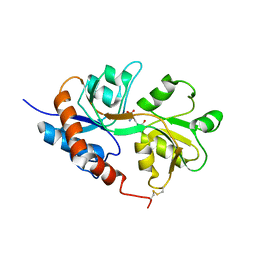 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
8TKP
 
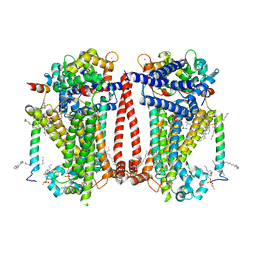 | | Structure of the C. elegans TMC-2 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, S, Jeong, H, Goehring, A, Posert, R, Gouaux, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structure of the Caenorhabditis elegans TMC-2 complex suggests roles of lipid-mediated subunit contacts in mechanosensory transduction.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3JAE
 
 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
3JAD
 
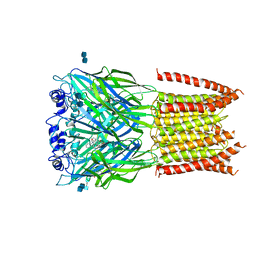 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, strychnine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, STRYCHNINE | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
3F4I
 
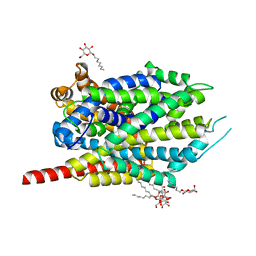 | | Crystal Structure of LeuT bound to L-selenomethionine and sodium | | Descriptor: | SELENOMETHIONINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3F4J
 
 | | Crystal structure of LeuT bound to glycine and sodium | | Descriptor: | GLYCINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
