8CRY
 
 | |
2O3H
 
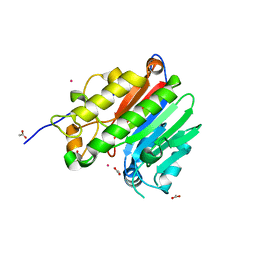 | | Crystal structure of the human C65A Ape | | Descriptor: | ACETATE ION, DNA-(apurinic or apyrimidinic site) lyase, SAMARIUM (III) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
1MML
 
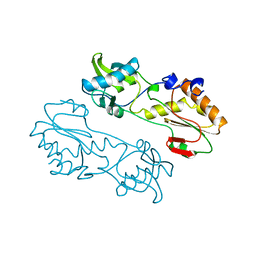 | | MECHANISTIC IMPLICATIONS FROM THE STRUCTURE OF A CATALYTIC FRAGMENT OF MMLV REVERSE TRANSCRIPTASE | | Descriptor: | MMLV REVERSE TRANSCRIPTASE | | Authors: | Georgiadis, M.M, Jessen, S.M, Ogata, C.M, Telesnitsky, A, Goff, S.P, Hendrickson, W.A. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase.
Structure, 3, 1995
|
|
4XO0
 
 | |
4XPC
 
 | |
4XPE
 
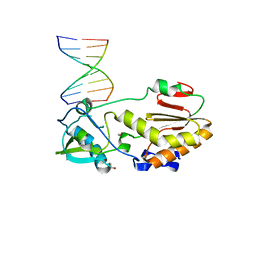 | |
4XNO
 
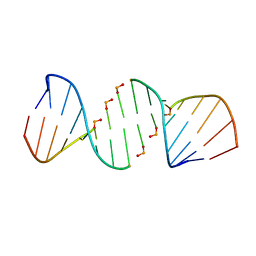 | | Crystal structure of 5'-CTTATPPPZZZATAAG | | Descriptor: | DNA (5'-D(*CP*TP*(BRU)P*AP*TP*(1WA)P*(1WA)P*(1WA)P*(1W5)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3') | | Authors: | Georgiadis, M.M, Singh, I. | | Deposit date: | 2015-01-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Structural basis for a six nucleotide genetic alphabet.
J. Am. Chem. Soc., 137, 2015
|
|
2O3C
 
 | | Crystal structure of zebrafish Ape | | Descriptor: | APEX nuclease 1, LEAD (II) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
9BVD
 
 | | Crystal structure of SRY HMG box bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*CP*TP*AP*GP*CP*AP*TP*TP*GP*TP*TP*TP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*AP*AP*AP*CP*AP*AP*TP*GP*CP*TP*AP*GP*TP*G)-3'), ... | | Authors: | Georgiadis, M.M. | | Deposit date: | 2024-05-20 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Role of nucleobase-specific interactions in the binding and bending of DNA by human male sex determination factor SRY.
J.Biol.Chem., 300, 2024
|
|
8CRZ
 
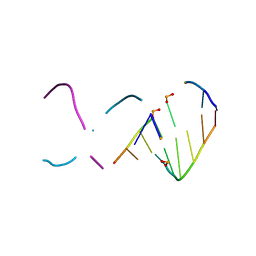 | | Crystal structure of alien DNA CTSZZPBSBSZPPBAG in a tetragonal lattice | | Descriptor: | DNA (5'-D(*CP*TP*(JSP)P*(1W5)P*(1W5)P*(1WA)P*(IGU)P*(JSP))-3'), DNA (5'-D(P*(IGU)P*(JSP)P*(1W5)P*(1WA)P*(1WA)P*(IGU)P*AP*G)-3') | | Authors: | Georgiadis, M.M. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Visualizing "Alternative Isoinformational Engineered" DNA in A- and B-Forms at High Resolution.
J.Am.Chem.Soc., 144, 2022
|
|
8CS0
 
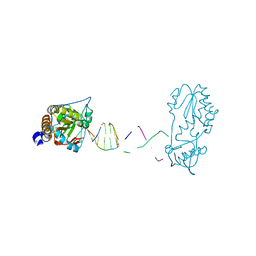 | |
1NIP
 
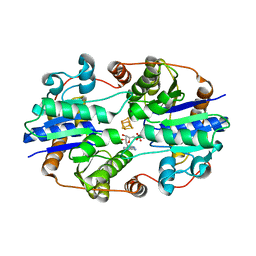 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii.
Science, 257, 1992
|
|
7UYO
 
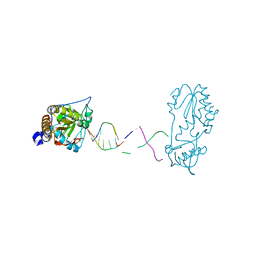 | |
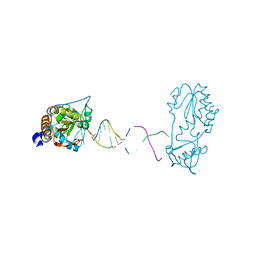
7UYN
 
 | |
7UYP
 
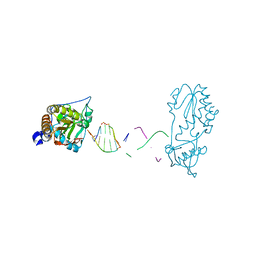 | |
7RAC
 
 | |
7RAB
 
 | |
6MIH
 
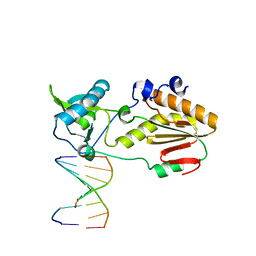 | | Crystal structure of host-guest complex with PC hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*(1WA)P*CP*(DB)P*T)-3'), DNA (5'-D(P*AP*(DS)P*GP*(1W5)P*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIK
 
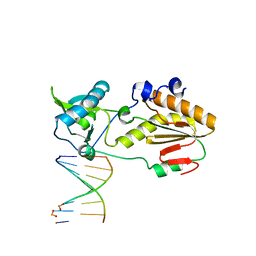 | | Crystal structure of host-guest complex with PP hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIG
 
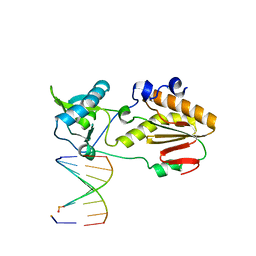 | | Crystal structure of host-guest complex with PB hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), Gag-Pol polyprotein | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MKM
 
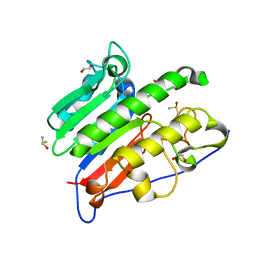 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MK3
 
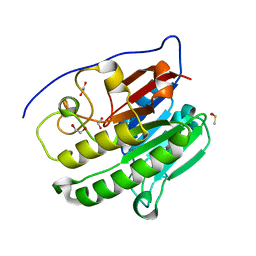 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MKO
 
 | |
6MKK
 
 | | Crystallographic solvent mapping analysis of DMSO/Mg bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
8DW1
 
 | | Crystal structure of a host-guest complex with 5'-CTTAGTTATAACTAAG-3' | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*GP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*CP*TP*AP*AP*G)-3'), reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2022-07-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Two distinct rotations of bithiazole DNA intercalation revealed by direct comparison of crystal structures of Co(III)•bleomycin A 2 and B 2 bound to duplex 5'-TAGTT sites.
Bioorg.Med.Chem., 77, 2023
|
|
