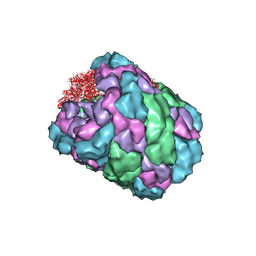
4BED
 
 | |
5LKI
 
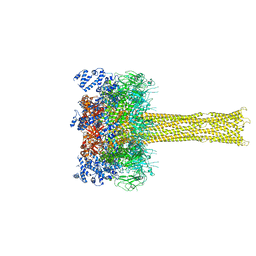 | | Cryo-EM structure of the Tc toxin TcdA1 in its pore state | | Descriptor: | TcdA1 | | Authors: | Gatsogiannis, C, Merino, F, Prumbaum, D, Roderer, D, Leidreiter, F, Meusch, D, Raunser, S. | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Membrane insertion of a Tc toxin in near-atomic detail.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5LKH
 
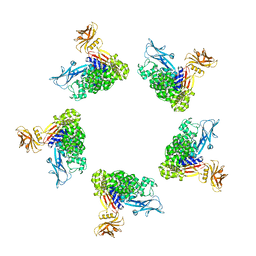 | | Cryo-EM structure of the Tc toxin TcdA1 in its pore state (obtained by flexible fitting) | | Descriptor: | TcdA1 | | Authors: | Gatsogiannis, C, Merino, F, Prumbaum, D, Roderer, D, Leidreiter, F, Meusch, D, Raunser, S. | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Membrane insertion of a Tc toxin in near-atomic detail.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6SFW
 
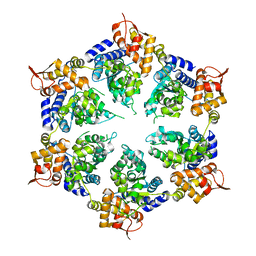 | |
6SFX
 
 | |
6H6E
 
 | | PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3) | | Descriptor: | TcdA1, TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
6H6G
 
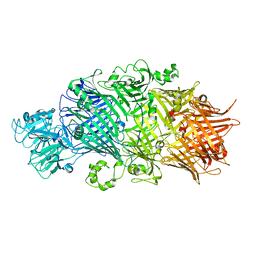 | | Crystal Structure of TcdB2-TccC3 without hypervariable C-terminal region | | Descriptor: | TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
6H6F
 
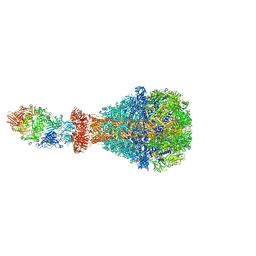 | | PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A | | Descriptor: | TcdA1, TcdB2,TccC3,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
8C0V
 
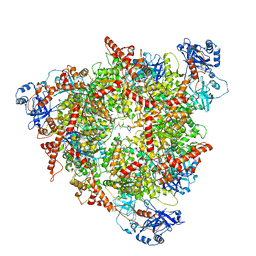 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in single seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8C0W
 
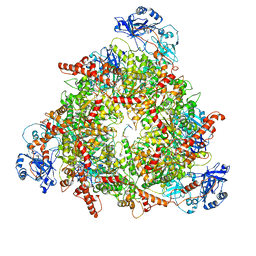 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
6XT9
 
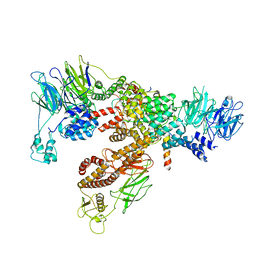 | | Subunits BBS 1,4,8,9,18 of the human BBSome complex | | Descriptor: | BBSome-interacting protein 1, Bardet-Biedl syndrome 1 protein, Bardet-Biedl syndrome 4 protein, ... | | Authors: | Klink, B.U, Raunser, S, Gatsogiannis, C. | | Deposit date: | 2020-01-15 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the human BBSome core complex.
Elife, 9, 2020
|
|
6XTB
 
 | |
6ZXK
 
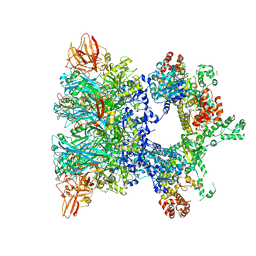 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state and PA7LF(2+1B) arrangement | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
6ZXL
 
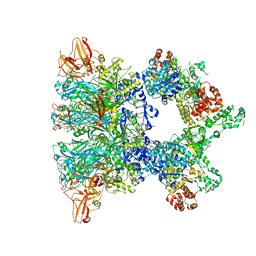 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state and PA7LF(2+1A) arrangement | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
6ZXJ
 
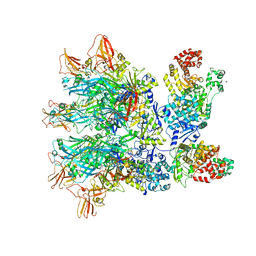 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state, in which the third lethal factor is masked out (PA7LF3-masked) | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
4O9Y
 
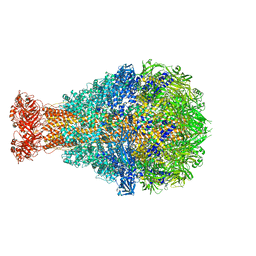 | | Crystal Structure of TcdA1 | | Descriptor: | TcdA1 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
4O9X
 
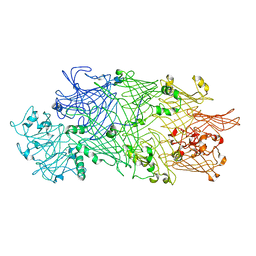 | | Crystal Structure of TcdB2-TccC3 | | Descriptor: | MERCURY (II) ION, TcdB2, TccC3 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
7QLA
 
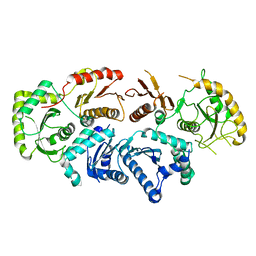 | | Structure of the Rab GEF complex Mon1-Ccz1 | | Descriptor: | Ccz1, Vacuolar fusion protein MON1 | | Authors: | Klink, B.U, Herrmann, E, Antoni, C, Langemeyer, L, Kiontke, S, Gatsogiannis, C, Ungermann, C, Raunser, S, Kuemmel, D. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of the Mon1-Ccz1 complex reveals molecular basis of membrane binding for Rab7 activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PTX
 
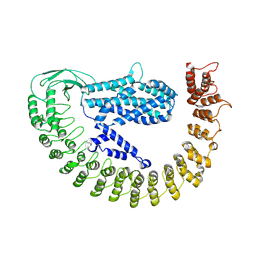 | | Alpha-latrocrustotoxin monomer | | Descriptor: | Alpha-latrocrustotoxin-Lt1a | | Authors: | Chen, M, Gatsogiannis, C. | | Deposit date: | 2021-09-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Molecular architecture of black widow spider neurotoxins.
Nat Commun, 12, 2021
|
|
7PTY
 
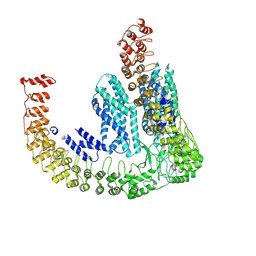 | | Delta-latroinsectotoxin dimer | | Descriptor: | Delta-latroinsectotoxin-Lt1a | | Authors: | Chen, M, Gatsogiannis, C. | | Deposit date: | 2021-09-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.63 Å) | | Cite: | Molecular architecture of black widow spider neurotoxins.
Nat Commun, 12, 2021
|
|
8ATD
 
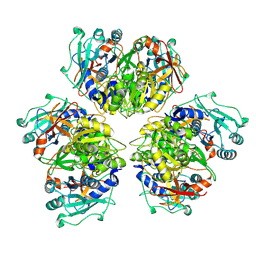 | | Wild type hexamer oxalyl-CoA synthetase (OCS) | | Descriptor: | Oxalate--CoA ligase | | Authors: | Lill, P, Burgi, J, Raunser, S, Wilmanns, M, Gatsogiannis, C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol.Chem., 404, 2023
|
|
6G1K
 
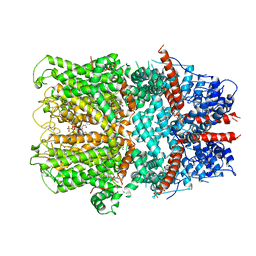 | | Electron cryo-microscopy structure of the canonical TRPC4 ion channel | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily c member 4a | | Authors: | Vinayagam, D, Mager, T, Apelbaum, A, Bothe, A, Merino, F, Hofnagel, O, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2018-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2018-08-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Electron cryo-microscopy structure of the canonical TRPC4 ion channel.
Elife, 7, 2018
|
|
7Q5P
 
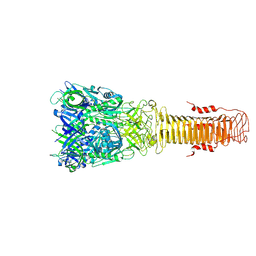 | | Structure of VgrG1 from Pseudomonas protegens. | | Descriptor: | Type VI secretion protein VgrG | | Authors: | Guenther, P, Quentin, D, Ahmad, S, Sachar, K, Gatsogiannis, C, Whitney, J.C, Raunser, S. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a bacterial Rhs effector exported by the type VI secretion system.
Plos Pathog., 18, 2022
|
|
7Q97
 
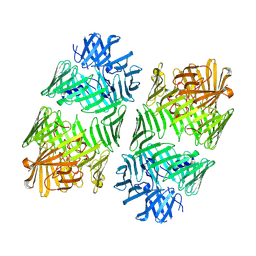 | | Structure of the bacterial type VI secretion system effector RhsA. | | Descriptor: | Rhs family protein | | Authors: | Guenther, P, Quentin, D, Ahmad, S, Sachar, K, Gatsogiannis, C, Whitney, J.C, Raunser, S. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a bacterial Rhs effector exported by the type VI secretion system.
Plos Pathog., 18, 2022
|
|
6R83
 
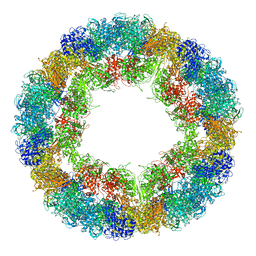 | | CryoEM structure and molecular model of squid hemocyanin (Todarodes pacificus , TpH) | | Descriptor: | Hemocyanin subunit 1 | | Authors: | Tanaka, Y, Kato, S, Stabrin, M, Raunser, S, Matsui, T, Gatsogiannis, C. | | Deposit date: | 2019-03-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM reveals the asymmetric assembly of squid hemocyanin.
Iucrj, 6, 2019
|
|
