5AN6
 
 | | Crystal structure of Thermotoga maritima Csm2 | | Descriptor: | CADMIUM ION, CRISPR-ASSOCIATED PROTEIN, CSM2 FAMILY | | Authors: | Gallo, G, Augusto, G, Rangel, G, Zelanis, A, Mori, M.A, Campos, C.B, Wurtele, M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-12-23 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural Basis for Dimer Formation of the Crispr-Associated Protein Csm2 of Thermotoga Maritima.
FEBS J., 283, 2016
|
|
7KWD
 
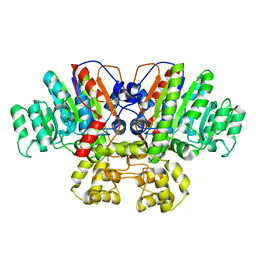 | | Crystal structure of Thermus thermophilus alkaline phosphatase | | Descriptor: | Alkaline phosphatase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Gallo, G, Coelho, C, Borges, B, Negri, N, Maiello, F, Hardy, L, Wurtele, M. | | Deposit date: | 2020-11-30 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamic cross correlation analysis of Thermus thermophilus alkaline phosphatase and determinants of thermostability.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6V6Y
 
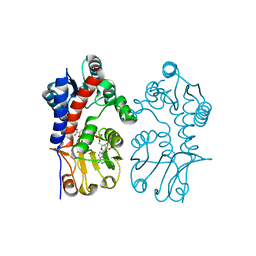 | | Crystal Structure of T. thermophilus methylenetetrahydrofolate dehydrogenase (MTHFD) | | Descriptor: | Bifunctional protein FolD, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maiello, F, Coelho, C, Gallo, G, Sucharski, F, Hardy, L, Wurtele, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15201569 Å) | | Cite: | Crystal structure of Thermus thermophilus methylenetetrahydrofolate dehydrogenase and determinants of thermostability.
Plos One, 15, 2020
|
|
5HYD
 
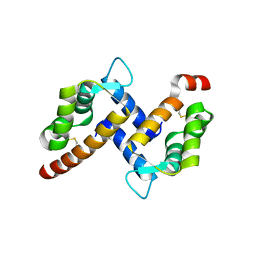 | |
6TI7
 
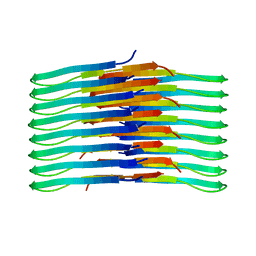 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TI6
 
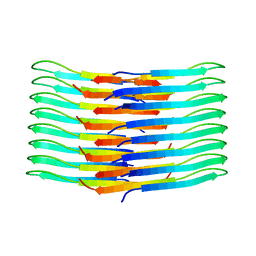 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
