9CCR
 
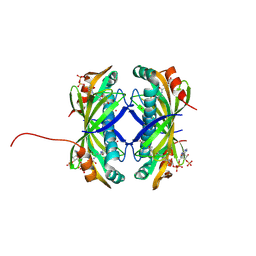 | | Crystal structure of the EspE7 thioesterase mutant R35A from the esperamicin biosynthetic pathway at 1.6 A | | Descriptor: | DODECYL-COA, POTASSIUM ION, Thioesterase | | Authors: | Miller, M.D, Hankore, E.D, Xu, W, Kosgei, A.J, Bhardwaj, M, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N. | | Deposit date: | 2024-06-23 | | Release date: | 2025-06-25 | | Last modified: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Studies on the Esperamicin Thioesterase and Progress toward Understanding Enediyne Core Biosynthesis.
J.Nat.Prod., 88, 2025
|
|
6VJV
 
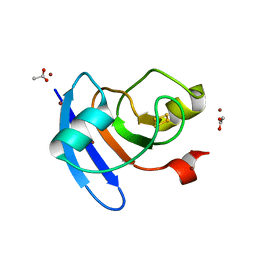 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
9N6T
 
 | |
7N7V
 
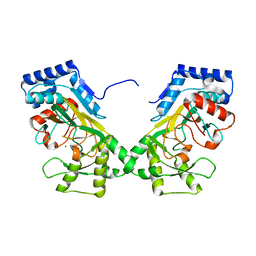 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
5HOQ
 
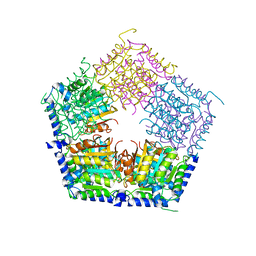 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
5INJ
 
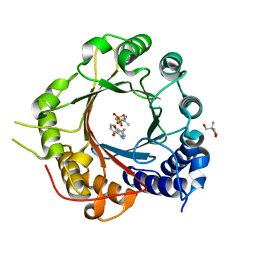 | | Crystal Structure of Prenyltransferase PriB Ternary Complex with L-Tryptophan and Dimethylallyl thiolodiphosphate (DMSPP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Prenyltransferase, ... | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
8JD7
 
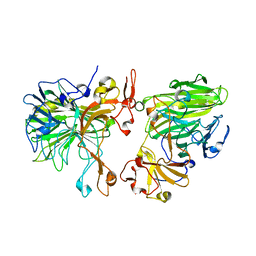 | |
8YA6
 
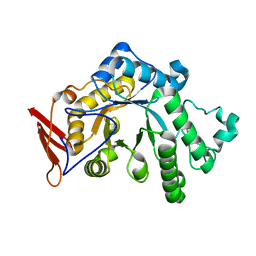 | | endo-1,3-fucanase Fun168A | | Descriptor: | endo-1,3-fucanase | | Authors: | Chen, G.N, Chang, Y.G. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Strucutre of endo-1,3-fucanase Fun168A at 1.92 Angstroms resolution.
To Be Published
|
|
8YA7
 
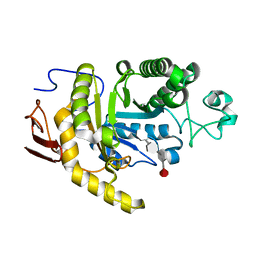 | | endo-1,3-fucanase Fun168A,complex with fucotetraose | | Descriptor: | alpha-L-fucopyranose-(1-3)-2,4-di-O-sulfo-alpha-L-fucopyranose-(1-3)-2-O-sulfo-alpha-L-fucopyranose-(1-3)-2-O-sulfo-alpha-L-fucopyranose, endo-1,3-fucanase | | Authors: | Chen, G.N, Chang, Y.G. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Strucutre of endo-1,3-fucanase Fun168A complex with fucotetrose at 1.99 Angstroms resolution.
To Be Published
|
|
8POE
 
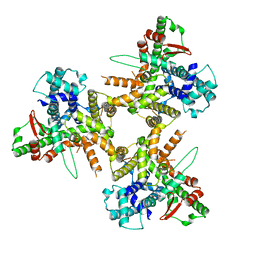 | | Structure of tissue-specific lipid scramblase ATG9B homotrimer, refined with C3 symmetry applied | | Descriptor: | Autophagy-related protein 9B | | Authors: | Chiduza, G.N, Pye, V.E, Tooze, S.A, Cherepanov, P. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | ATG9B is a tissue-specific homotrimeric lipid scramblase that can compensate for ATG9A.
Autophagy, 20, 2024
|
|
5K9M
 
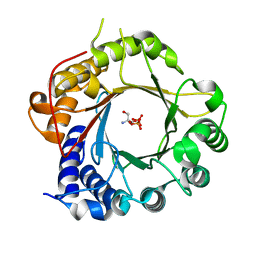 | | Crystal Structure of PriB Binary Complex with Product Diphosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYROPHOSPHATE 2-, PriB Prenyltransferase | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-06-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5JXM
 
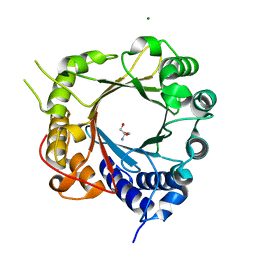 | | Crystal Structure of Prenyltransferase PriB Apo Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PriB | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
6AX7
 
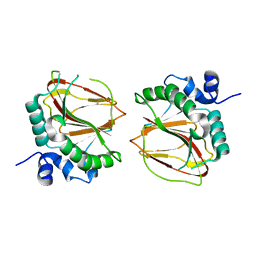 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
8QQH
 
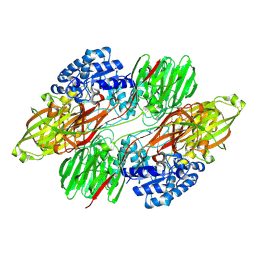 | |
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6N04
 
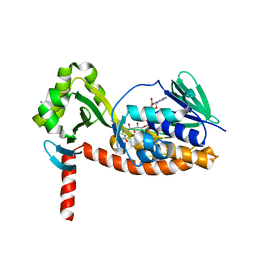 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
7UX8
 
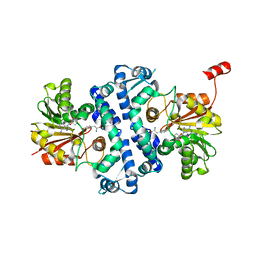 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH and L-Tyrosine bound at 1.4 A resolution (P212121 - form II) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, TYROSINE, ... | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
7UX7
 
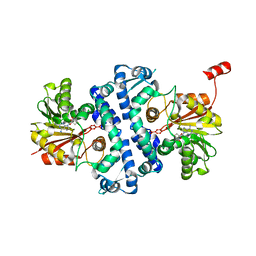 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH bound at 1.2 A resolution (P212121 - form II) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
7UX6
 
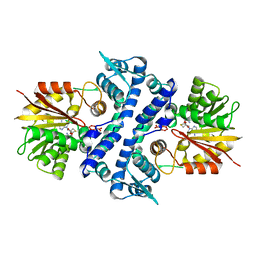 | | Crystal structure of MfnG, an L- and D-tyrosine O-methyltransferase from the marformycin biosynthesis pathway of Streptomyces drozdowiczii, with SAH bound at 1.35 A resolution (P212121 - form I) | | Descriptor: | MfnG, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Miller, M.D, Wu, K.-L, Xu, W, Xiao, H, Philips Jr, G.N. | | Deposit date: | 2022-05-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.
Protein Sci., 31, 2022
|
|
5JR3
 
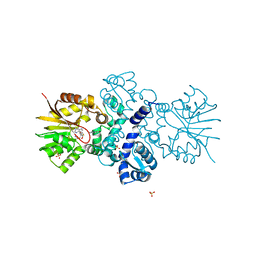 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Johnson, B.R, Huber, T.D, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 4-methylumbelliferone (to be published)
To Be Published
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UC4
 
 | |
6QI5
 
 | | Near Atomic Structure of an Atadenovirus Shows a possible gene duplication event and Intergenera Variations in Cementing Proteins | | Descriptor: | Hexon protein, PIIIa, Penton protein, ... | | Authors: | Condezo, G.N, Marabini, R, Gomez-Blanco, J, SanMartin, C. | | Deposit date: | 2019-01-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins.
Sci Adv, 7, 2021
|
|
