3RUB
 
 | | CRYSTAL STRUCTURE OF THE UNACTIVATED FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM TOBACCO REFINED AT 2.0-ANGSTROMS RESOLUTION | | Descriptor: | ASPARAGINE, RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, FORM III, ... | | Authors: | Schreuder, H, Cascio, D, Curmi, P.M.G, Chapman, M.S, Suh, S.W, Eisenberg, D.S. | | Deposit date: | 1990-05-25 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution.
J.Biol.Chem., 267, 1992
|
|
1XF6
 
 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | Descriptor: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | Authors: | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | Deposit date: | 2004-09-14 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
1XG0
 
 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | Descriptor: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | Authors: | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
1MGQ
 
 | |
3OQS
 
 | | Crystal structure of importin-alpha bound to a CLIC4 NLS peptide | | Descriptor: | Importin subunit alpha-2, peptide of Chloride intracellular channel protein 4 | | Authors: | Mynott, A.V, Brown, L.J, Harrop, S.J, Curmi, P.M.G. | | Deposit date: | 2010-09-03 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of importin-alpha bound to a peptide bearing the nuclear localisation signal from chloride intracellular channel protein 4
Febs J., 278, 2011
|
|
2AHE
 
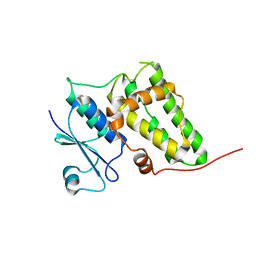 | | Crystal structure of a soluble form of CLIC4. intercellular chloride ion channel | | Descriptor: | Chloride intracellular channel protein 4 | | Authors: | Littler, D.R, Assaad, N.N, Harrop, S.J, Brown, L.J, Pankhurst, G.J, Luciani, P, Aguilar, M.-I, Mazzanti, M, Berryman, M.A, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4.
FEBS J., 272, 2005
|
|
3KJY
 
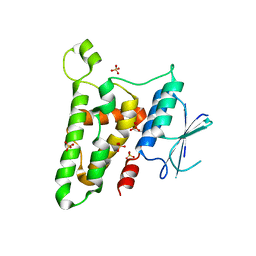 | | Crystal structure of reduced HOMO SAPIENS CLIC3 | | Descriptor: | Chloride intracellular channel protein 3, SULFATE ION | | Authors: | Littler, D.R, Curmi, P.M.G, Breit, S.N, Perrakis, A. | | Deposit date: | 2009-11-04 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of human CLIC3 at 2 A resolution
Proteins, 78, 2010
|
|
3BW1
 
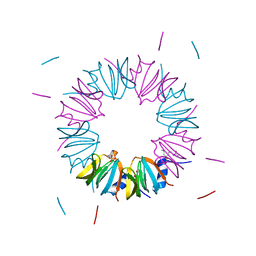 | | Crystal structure of homomeric yeast Lsm3 exhibiting novel octameric ring organisation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, SULFATE ION, U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Naidoo, N, Harrop, S.J, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2008-01-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Lsm3 octamer from Saccharomyces cerevisiae: implications for Lsm ring organisation and recruitment
J.Mol.Biol., 377, 2008
|
|
8EL4
 
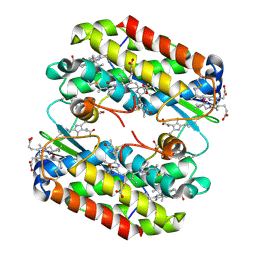 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in a tight interface filament | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
8EL3
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in a loose interface filament | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
8EL6
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 with an altered helix hA/hY conformation | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
8EL5
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in an alternating tight to loose interface filament | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, GLYCEROL, PHYCOERYTHROBILIN, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
1JRR
 
 | | HUMAN PLASMINOGEN ACTIVATOR INHIBITOR-2.[LOOP (66-98) DELETIONMUTANT] COMPLEXED WITH PEPTIDE MIMIckING THE REACTIVE CENTER LOOP | | Descriptor: | BETA-MERCAPTOETHANOL, PLASMINOGEN ACTIVATOR INHIBITOR-2 | | Authors: | Jankova, L, Harrop, S.J, Saunders, D.N, Andrews, J.L, Bertram, K.C, Gould, A.R, Baker, M.S, Curmi, P.M.G. | | Deposit date: | 2001-08-15 | | Release date: | 2001-09-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF THE COMPLEX OF PLASMINOGEN ACTIVATOR INHIBITOR 2 WITH A PEPTIDE MIMICKING THE REACTIVE CENTER LOOP
J.Biol.Chem., 276, 2001
|
|
1N9S
 
 | | Crystal structure of yeast SmF in spacegroup P43212 | | Descriptor: | Small nuclear ribonucleoprotein F | | Authors: | Collins, B.M, Cubeddu, L, Naidoo, N, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2002-11-26 | | Release date: | 2002-12-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Homomeric ring assemblies of eukaryotic Sm proteins have affinity for
both RNA and DNA: Crystal structure of an oligomeric complex of yeast
SmF
J.Biol.Chem., 278, 2003
|
|
1N9R
 
 | | Crystal structure of a heptameric ring complex of yeast SmF in spacegroup P4122 | | Descriptor: | Small nuclear ribonucleoprotein F | | Authors: | Collins, B.M, Cubeddu, L, Naidoo, N, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2002-11-26 | | Release date: | 2002-12-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homomeric ring assemblies of eukaryotic Sm proteins have affinity for
both RNA and DNA: Crystal structure of an oligomeric complex of yeast
SmF
J.Biol.Chem., 278, 2003
|
|
4K0G
 
 | | Crystal structure of human CLIC1 C24S mutant | | Descriptor: | ACETATE ION, CALCIUM ION, Chloride intracellular channel protein 1 | | Authors: | Phang, J.M, Harrop, S.J, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2013-04-03 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure analysis of CLIC1 C24 mutants
To be Published
|
|
3EY8
 
 | | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, SULFATE ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1
To be Published
|
|
3EY7
 
 | | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, CALCIUM ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1
To be Published
|
|
4JZQ
 
 | |
4K0N
 
 | | Crystal structure of human CLIC1 C24A mutant | | Descriptor: | Chloride intracellular channel protein 1, MALONATE ION | | Authors: | Phang, J.M, Harrop, S.J, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure analysis of CLIC1 C24 mutants
To be Published
|
|
7SSF
 
 | | Light harvesting phycobiliprotein HaPE560 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DiCys-(15,16)-Dihydrobiliverdin, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-10 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii
Commun Biol, 2023
|
|
7SUT
 
 | | Light harvesting phycobiliprotein HaPE645 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | (15,16)-DIHYDROBILIVERDIN (SINGLY LINKED), 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
7T7U
 
 | | Light Harvesting complex phycocyanin PC 630, from the cryptophyte Chroomonas sp. M1627 | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, GLYCEROL, PHYCOCYANOBILIN, ... | | Authors: | Michie, K.A, Harrop, S.J, Rathbone, H.W, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2021-12-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
7T8S
 
 | | Light Harvesting complex phycoerythrin PE 566, from the cryptophyte Cryptomonas pyrenoidifera | | Descriptor: | Bilin 584 (doubly linked), Bilin 584 (single linked), Bilin 618 (single linked), ... | | Authors: | Michie, K.A, Harrop, S.J, Rathbone, H.W, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2021-12-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
2YV9
 
 | |
