1YPI
 
 | |
1L02
 
 | |
1L09
 
 | |
1L06
 
 | |
1L15
 
 | |
1L13
 
 | |
3F69
 
 | | Crystal structure of the Mycobacterium tuberculosis PknB mutant kinase domain in complex with KT5720 | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase pknB, hexyl (5S,6R,8R)-6-hydroxy-5-methyl-13-oxo-5,6,7,8-tetrahydro-13H-5,8-epoxy-4b,8a,14-triazadibenzo[b,h]cycloocta[1,2,3,4-jkl]c yclopenta[e]-as-indacene-6-carboxylate | | Authors: | Alber, T, Mieczkowski, C.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase.
Embo J., 27, 2008
|
|
3NBU
 
 | | Crystal structure of pGI glucosephosphate isomerase | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase | | Authors: | Alber, T, Zubieta, C, Totir, M, May, A, Echols, N. | | Deposit date: | 2010-06-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
3UQC
 
 | |
4IMY
 
 | |
1L12
 
 | |
1L03
 
 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | Deposit date: | 1988-02-05 | | Release date: | 1988-04-16 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
1L07
 
 | |
1L08
 
 | |
1L04
 
 | |
1L05
 
 | |
1L11
 
 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | Deposit date: | 1988-02-05 | | Release date: | 1988-04-16 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
1L14
 
 | |
3K7Z
 
 | |
2I6F
 
 | | Receiver domain from Myxococcus xanthus social motility protein FrzS | | Descriptor: | CHLORIDE ION, Response regulator FrzS | | Authors: | Echols, N, Fraser, J, Merlie, J, Zusman, D, Alber, T. | | Deposit date: | 2006-08-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
1TXO
 
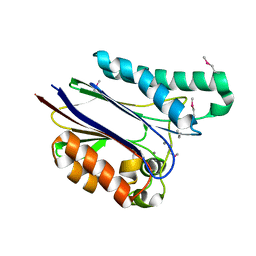 | | Crystal structure of the Mycobacterium tuberculosis serine/threonine phosphatase PstP/Ppp at 1.95 A. | | Descriptor: | MANGANESE (II) ION, Putative Bacterial Enzyme | | Authors: | Pullen, K.E, Ng, H.L, Sung, P.Y, Good, M.C, Smith, S.M, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-05 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An Alternate Conformation and a Third Metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr Protein Phosphatase.
Structure, 12, 2004
|
|
1ZIL
 
 | |
1ZIM
 
 | |
1ZIK
 
 | |
1ZII
 
 | |
