1Z9D
 
 | | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes | | Descriptor: | SULFATE ION, uridylate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-04-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative uridylate kinase (UMP-kinase) from Streptococcus pyogenes
To be Published
|
|
2A8X
 
 | | Crystal Structure of Lipoamide Dehydrogenase from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rajashankar, K.R, Bryk, R, Kniewel, R, Buglino, J.A, Nathan, C.F, Lima, C.D. | | Deposit date: | 2005-07-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis
J.Biol.Chem., 280, 2005
|
|
2AP9
 
 | | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551 | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, acetylglutamate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-15 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551
To be Published
|
|
2AR0
 
 | |
1L9G
 
 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
1SZQ
 
 | | Crystal Structure of 2-methylcitrate dehydratase | | Descriptor: | 2-methylcitrate dehydratase | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-06 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 2-methylcitrate dehydratase
To be Published
|
|
1T35
 
 | | CRYSTAL STRUCTURE OF A HYPOTHETICAL PROTEIN YVDD- A PUTATIVE LYSINE DECARBOXYLASE | | Descriptor: | HYPOTHETICAL PROTEIN YVDD, Putative Lysine Decarboxylase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structure of a Hypothetical Protein Yvdd - Putative Lysine Decarboxylase
To be Published
|
|
1TIK
 
 | | CRYSTAL STRUCTURE OF ACYL CARRIER PROTEIN PHOSPHODIESTERASE | | Descriptor: | Acyl carrier protein phosphodiesterase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ACYL CARRIER PROTEIN PHOSPHODIESTERASE
To be Published
|
|
1T3U
 
 | | Unknown conserved bacterial protein from Pseudomonas aeruginosa PAO1 | | Descriptor: | conserved hypothetical protein | | Authors: | Rajashankar, K.R, Kneiwel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-04 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a conserved hypothetical protein Pseudomonas aeruginosa PA01
To be Published
|
|
1TLT
 
 | | Crystal Structure of a Putative Oxidoreductase (VIRULENCE FACTOR mviM HOMOLOG) | | Descriptor: | PUTATIVE OXIDOREDUCTASE (VIRULENCE FACTOR mviM HOMOLOG), SULFATE ION | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Putative Oxidoreductase (VIRULENCE FACTOR mviM HOMOLOG)
To be Published
|
|
1TWI
 
 | | Crystal structure of Diaminopimelate Decarboxylase from m. jannaschii in co-complex with L-lysine | | Descriptor: | Diaminopimelate decarboxylase, LYSINE, MAGNESIUM ION, ... | | Authors: | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M.G, He, G, De Lencastre, H, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-01 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor
Structure, 10, 2002
|
|
1TO6
 
 | | Glycerate kinase from Neisseria meningitidis (serogroup A) | | Descriptor: | Glycerate kinase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-13 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycerate kinase from Neisseria meningitidis (serogroup A)
To be Published
|
|
1TQ8
 
 | | Crystal Structure of protein Rv1636 from Mycobacterium tuberculosis H37Rv | | Descriptor: | hypothetical protein Rv1636 | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hypothetical protein Rv1636 from Mycobacterium tuberculosis H37Rv
To be Published
|
|
1TVF
 
 | | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus | | Descriptor: | SULFATE ION, UNKNOWN LIGAND, penicillin binding protein 4 | | Authors: | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-29 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus
To be Published
|
|
1XRH
 
 | |
1Y8C
 
 | | Crystal structure of a S-adenosylmethionine-dependent methyltransferase from Clostridium acetobutylicum ATCC 824 | | Descriptor: | S-adenosylmethionine-dependent methyltransferase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-11 | | Release date: | 2004-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a S-adenosylmethionine-dependent methyltransferase from Clostridium acetobutylicum ATCC 824
To be Published
|
|
1Y23
 
 | |
1YCO
 
 | | Crystal structure of a branched-chain phosphotransacylase from Enterococcus faecalis V583 | | Descriptor: | PHOSPHATE ION, branched-chain phosphotransacylase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-22 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a branched-chain phosphotransacylase from Enterococcus faecalis V583
To be Published
|
|
1TK9
 
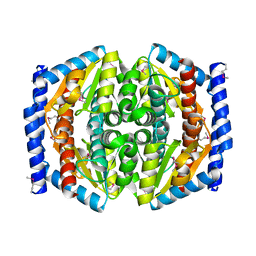 | | Crystal Structure of Phosphoheptose isomerase 1 | | Descriptor: | Phosphoheptose isomerase 1 | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two putative phosphoheptose isomerases.
Proteins, 63, 2006
|
|
2PMV
 
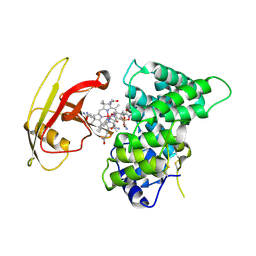 | | Crystal Structure of Human Intrinsic Factor- Cobalamin Complex at 2.6 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COBALAMIN, Gastric intrinsic factor | | Authors: | Mathews, F.S, Gordon, M.M, Chen, Z, Rajashankar, K.R, Ealick, S.E, Alpers, D.H, Sukumar, N. | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human intrinsic factor: Cobalamin complex at 2.6-A resolution
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6CIH
 
 | | Crystal structure of a group II intron lariat in the post-catalytic state | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*AP*C*-3'), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
6D9Z
 
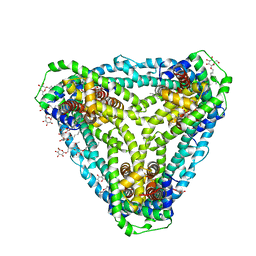 | | Structure of CysZ, a sulfate permease from Pseudomonas Denitrificans | | Descriptor: | Sulfate transporter CysZ, octyl beta-D-glucopyranoside | | Authors: | Sanghai, Z.A, Clarke, O.B, Liu, Q, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Mancia, F. | | Deposit date: | 2018-04-30 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4021318 Å) | | Cite: | Structure-based analysis of CysZ-mediated cellular uptake of sulfate.
Elife, 7, 2018
|
|
6D79
 
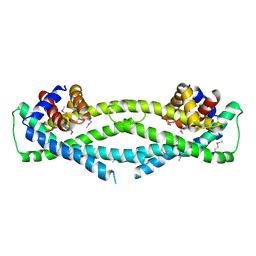 | | Structure of CysZ, a sulfate permease from Pseudomonas Fragi | | Descriptor: | Sulfate transporter CysZ | | Authors: | Sanghai, Z.A, Liu, Q, Clarke, O.B, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Mancia, F. | | Deposit date: | 2018-04-24 | | Release date: | 2018-05-16 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structure-based analysis of CysZ-mediated cellular uptake of sulfate.
Elife, 7, 2018
|
|
1E6F
 
 | | Human MIR-receptor, repeat 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Rajashankar, K.R, Dauter, M, Dauter, Z, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4ENC
 
 | | Crystal structure of fluoride riboswitch | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
