1ATG
 
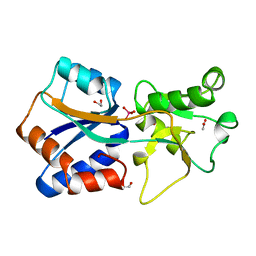 | | AZOTOBACTER VINELANDII PERIPLASMIC MOLYBDATE-BINDING PROTEIN | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PERIPLASMIC MOLYBDATE-BINDING PROTEIN, ... | | Authors: | Lawson, D.M, Pau, R.N, Williams, C.E.M, Mitchenall, L.A. | | Deposit date: | 1997-08-14 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA.
Structure, 6, 1998
|
|
1THT
 
 | | STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI | | Descriptor: | THIOESTERASE | | Authors: | Lawson, D.M, Derewenda, U, Serre, L, Ferri, S, Szitter, R, Wei, Y, Meighen, E.A, Derewenda, Z.S. | | Deposit date: | 1994-04-19 | | Release date: | 1995-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi.
Biochemistry, 33, 1994
|
|
1E84
 
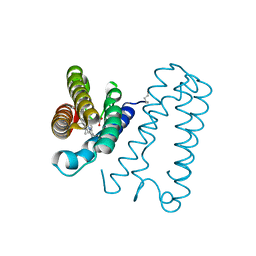 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E86
 
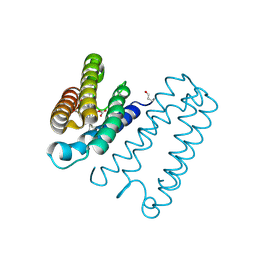 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with CO bound to distal side of heme | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E85
 
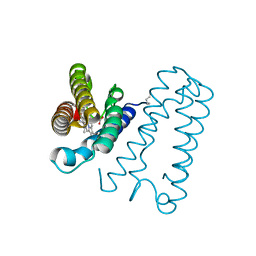 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with NO bound to proximal side of heme | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase.
Embo J., 19, 2000
|
|
1E83
 
 | | Cytochrome c' from Alcaligenes xylosoxidans - oxidized structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
8PFC
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PFD
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the von Willebrand Factor Type A domain of the proteasomal ubiquitin receptor Rpn10 from Arabidopsis thaliana | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7OL9
 
 | |
7NFU
 
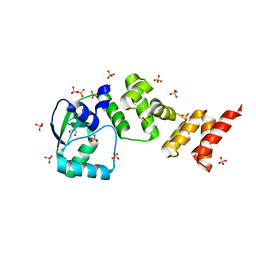 | | Crystal structure of C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | GLYCEROL, Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
7NG0
 
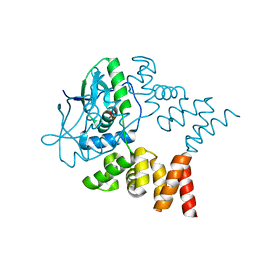 | | Crystal structure of N- and C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
4CKK
 
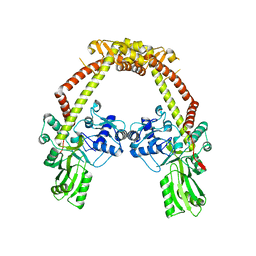 | | Apo structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit | | Descriptor: | DNA GYRASE SUBUNIT A | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
4CKL
 
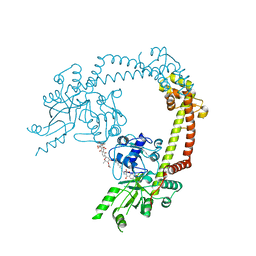 | | Structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit with simocyclinone D8 bound | | Descriptor: | DNA GYRASE SUBUNIT A, SIMOCYCLINONE D8 | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
6F8J
 
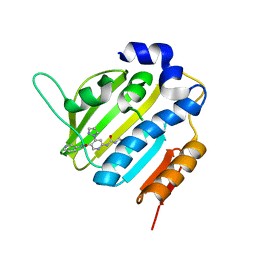 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F96
 
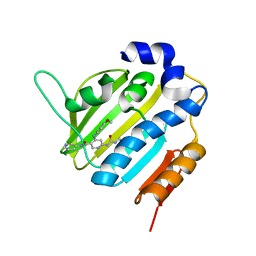 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F9Q
 
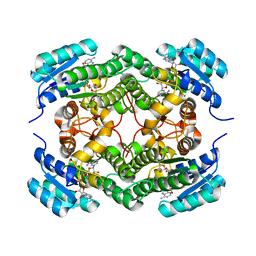 | | Binary complex of a 7S-cis-cis-nepetalactol cyclase from Nepeta mussinii with NAD+ | | Descriptor: | 7S-cis-cis-nepetalactol cyclase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lichman, B.R, Kamileen, M.O, Titchiner, G, Saalbach, G, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
9FOY
 
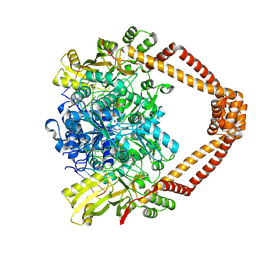 | | Ternary complex of a Mycobacterium tuberculosis DNA gyrase core fusion with DNA and the inhibitor AMK32b | | Descriptor: | (1~{R})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, (1~{S})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), ... | | Authors: | Kokot, M, Hrast, M, Feng, L, Mitchenall, L.A, Lawson, D.M, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2024-06-12 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Aspects of Mycobacterium tuberculosis DNA Gyrase Targeted by Novel Bacterial Topoisomerase Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
2UYN
 
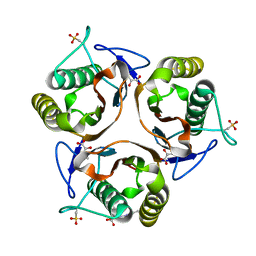 | | Crystal structure of E. coli TdcF with bound 2-ketobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2V09
 
 | | SENS161-164DSSN mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Burrell, M.R, Just, V.J, Bowater, L, Fairhurst, S.A, Requena, L, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxalate Decarboxylase and Oxalate Oxidase Activities Can be Interchanged with a Specificity Switch of Up to 282 000 by Mutating an Active Site Lid.
Biochemistry, 46, 2007
|
|
2UYJ
 
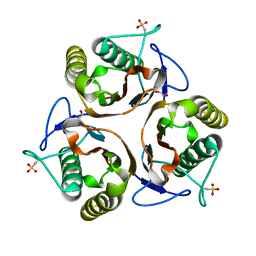 | | Crystal structure of E. coli TdcF with bound ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2UYA
 
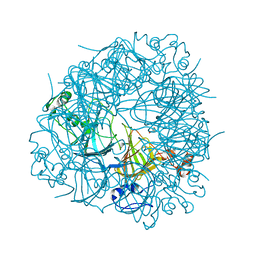 | | DEL162-163 mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UYB
 
 | | S161A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UYP
 
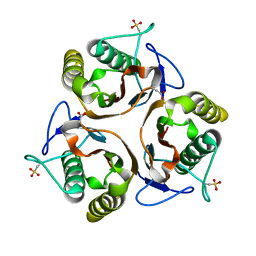 | | Crystal structure of E. coli TdcF with bound propionate | | Descriptor: | PROPANOIC ACID, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2UYK
 
 | | Crystal structure of E. coli TdcF with bound serine | | Descriptor: | PROTEIN TDCF, SERINE | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
5CQ1
 
 | | Disproportionating enzyme 1 from Arabidopsis - cycloamylose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
