6PPT
 
 | |
6PQ2
 
 | |
2L5F
 
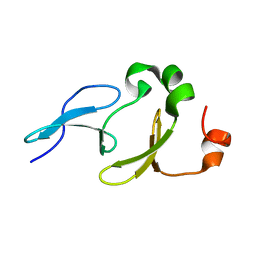 | | Solution structure of the tandem WW domains from HYPA/FBP11 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interaction with polyglutamine expanded huntingtin alters cellular distribution and RNA processing of huntingtin yeast two-hybrid protein A (HYPA)
To be Published
|
|
1LNQ
 
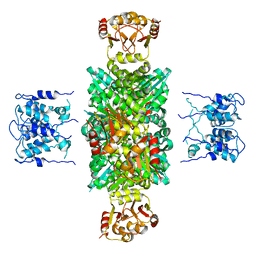 | | CRYSTAL STRUCTURE OF MTHK AT 3.3 A | | Descriptor: | CALCIUM ION, POTASSIUM CHANNEL RELATED PROTEIN | | Authors: | Jiang, Y, Lee, A, Chen, J, Cadene, M, Chait, B.T, Mackinnon, R. | | Deposit date: | 2002-05-03 | | Release date: | 2002-06-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | CRYSTAL STRUCTURE AND MECHANISM OF A CALCIUM-GATED POTASSIUM CHANNEL
Nature, 417, 2002
|
|
6PQM
 
 | |
6PRQ
 
 | |
6PRJ
 
 | |
6PRP
 
 | |
6PQE
 
 | |
6PRI
 
 | |
6PSI
 
 | |
5X74
 
 | | The crystal Structure PDE delta in complex with compound (R, R)-1g | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-[2-[4-[(2R)-2-(2-fluorophenyl)-4-oxidanylidene-1,2-dihydroquinazolin-3-yl]piperidin-1-yl]ethyl]-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5X73
 
 | | The crystal Structure PDE delta in complex with R-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5EFX
 
 | | Crystal structure of Rho GTPase regulator | | Descriptor: | Rho guanine nucleotide exchange factor 2 | | Authors: | Jiang, Y, Ouyang, S, Liu, Z.J. | | Deposit date: | 2015-10-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structure of hGEF-H1 PH domain provides insight into incapability in phosphoinositide binding
Biochem.Biophys.Res.Commun., 471, 2016
|
|
1ID1
 
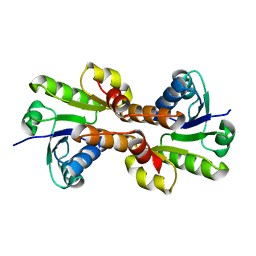 | | CRYSTAL STRUCTURE OF THE RCK DOMAIN FROM E.COLI POTASSIUM CHANNEL | | Descriptor: | PUTATIVE POTASSIUM CHANNEL PROTEIN | | Authors: | Jiang, Y, Pico, A, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel.
Neuron, 29, 2001
|
|
1ORS
 
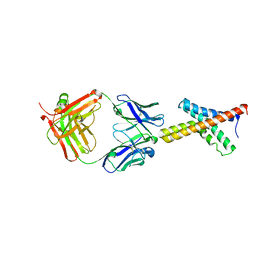 | | X-ray structure of the KvAP potassium channel voltage sensor in complex with an Fab | | Descriptor: | 33H1 Fab heavy chain, 33H1 Fab light chain, potassium channel | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
1ORQ
 
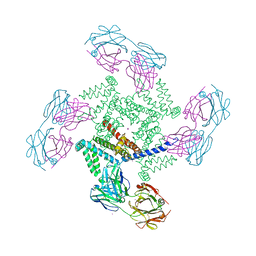 | | X-ray structure of a voltage-dependent potassium channel in complex with an Fab | | Descriptor: | 6E1 Fab heavy chain, 6E1 Fab light chain, CADMIUM ION, ... | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
1TFE
 
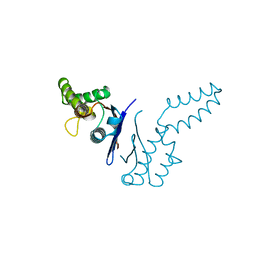 | | DIMERIZATION DOMAIN OF EF-TS FROM T. THERMOPHILUS | | Descriptor: | ELONGATION FACTOR TS | | Authors: | Jiang, Y, Nock, S, Nesper, M, Sprinzl, M, Sigler, P.B. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and importance of the dimerization domain in elongation factor Ts from Thermus thermophilus.
Biochemistry, 35, 1996
|
|
3QWW
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the methyltransferase inhibitor sinefungin | | Descriptor: | SET and MYND domain-containing protein 2, SINEFUNGIN, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
3QWV
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the cofactor product AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 2, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
2LJ1
 
 | | The third SH3 domain of R85FL with ataxin-7 PRR | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
2LJ0
 
 | | The third SH3 domain of R85FL | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
8TEL
 
 | |
8TEH
 
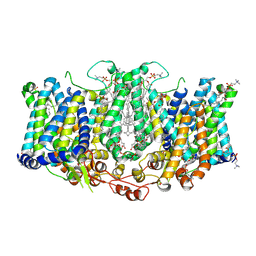 | | Cryo-EM structure of Arabidopsis thaliana Bor1 in lipid nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Boron transporter 1 | | Authors: | Jiang, Y, Jiang, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure of borate transporter Bor1 reveals a novel auto-inhibition mechanism for
the SLC4 family
To Be Published
|
|
8TEG
 
 | |
