1QM4
 
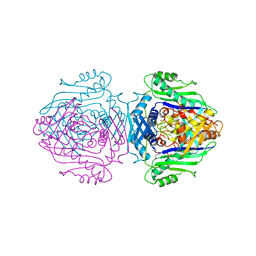 | | Methionine Adenosyltransferase Complexed with a L-Methionine Analogue | | Descriptor: | L-2-AMINO-4-METHOXY-CIS-BUT-3-ENOIC ACID, MAGNESIUM ION, METHIONINE ADENOSYLTRANSFERASE, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Crystal Structure of Tetrameric Methionine Adenosyltransferase from Rat Liver Reveals the Methionine-Binding Site
J.Mol.Biol., 300, 2000
|
|
2XAO
 
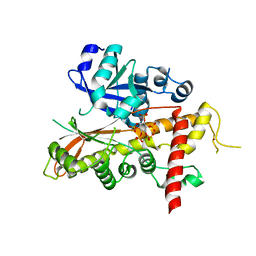 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with IP5 | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, ZINC ION | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1UMY
 
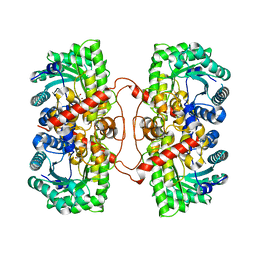 | | BHMT from rat liver | | Descriptor: | BETA-MERCAPTOETHANOL, Betaine--homocysteine S-methyltransferase 1, ZINC ION | | Authors: | Gonzalez, B, Pajares, M.A, Sanz-Aparicio, J. | | Deposit date: | 2003-09-02 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat liver betaine homocysteine s-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding.
J. Mol. Biol., 338, 2004
|
|
1O90
 
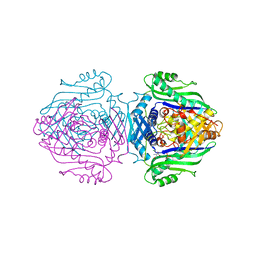 | | Methionine Adenosyltransferase complexed with a L-methionine analogue | | Descriptor: | (2S,4S)-2-AMINO-4,5-EPOXIPENTANOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1O92
 
 | | Methionine Adenosyltransferase complexed with ADP and a L-methionine analogue | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-2-AMINO-4-METHOXY-CIS-BUT-3-ENOIC ACID, MAGNESIUM ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1O9T
 
 | | Methionine adenosyltransferase complexed with both substrates ATP and methionine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, METHIONINE, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1O93
 
 | | Methionine Adenosyltransferase complexed with ATP and a L-methionine analogue | | Descriptor: | (2S,4S)-2-AMINO-4,5-EPOXIPENTANOIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1W2F
 
 | | Human Inositol (1,4,5)-trisphosphate 3-kinase substituted with selenomethionine | | Descriptor: | INOSITOL-TRISPHOSPHATE 3-KINASE A, SULFATE ION | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1W2C
 
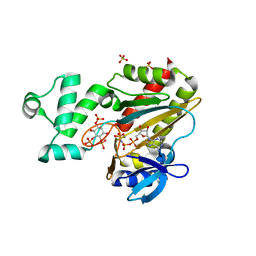 | | Human Inositol (1,4,5) trisphosphate 3-kinase complexed with Mn2+/AMPPNP/Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, MANGANESE (II) ION, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
2XAL
 
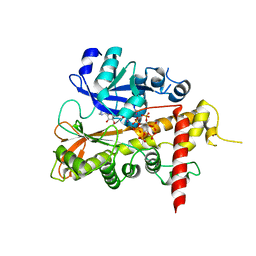 | | Lead derivative of Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with ADP and IP6. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XAM
 
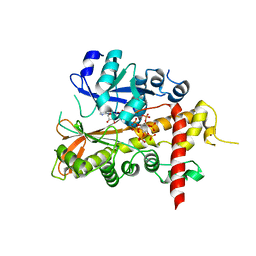 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with ADP and IP6. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XAN
 
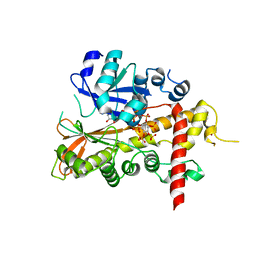 | | inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with AMP PNP and IP5 | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, MAGNESIUM ION, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XAR
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with IP6. | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ZINC ION | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1W2D
 
 | | Human Inositol (1,4,5)-trisphosphate 3-kinase complexed with Mn2+/ADP/Ins(1,3,4,5)P4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
6WKK
 
 | | Phage G gp27 major capsid proteins and gp26 decoration proteins | | Descriptor: | Gp26 capsid decoration protein, Gp27 major capsid protein | | Authors: | Monroe, L, Gonzalez, B, Jiang, W, Kihara, D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Phage G Structure at 6.1 angstrom Resolution, Condensed DNA, and Host Identity Revision to a Lysinibacillus.
J.Mol.Biol., 432, 2020
|
|
5MWL
 
 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH ATP and IP5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, MAGNESIUM ION, ... | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
7AUU
 
 | |
7AUT
 
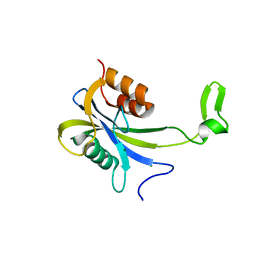 | |
7AUN
 
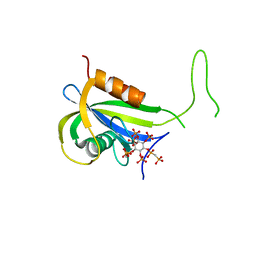 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PCP-InsP8 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, MAGNESIUM ION, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(hydroxyphosphoryl)methanediyl]}bis(phosphonic acid) | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUO
 
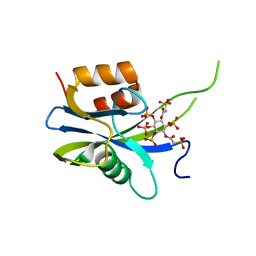 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PA-InsP8 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(2-oxoethane-2,1-diyl)]}bis(phosphonic acid) | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUP
 
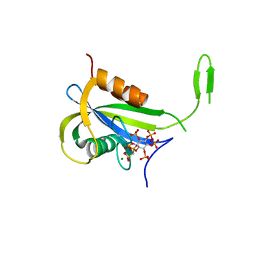 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PCP-InsP7 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, MAGNESIUM ION, [oxidanyl-[(2~{S},3~{S},5~{R},6~{S})-2,3,4,5,6-pentaphosphonooxycyclohexyl]oxy-phosphoryl]methylphosphonic acid | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUL
 
 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with 5-InsP7 in presence of Mg | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ACETATE ION, Diphosphoinositol polyphosphate phosphohydrolase DDP1, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUR
 
 | |
7AUK
 
 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with 5-InsP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Diphosphoinositol polyphosphate phosphohydrolase DDP1 | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
7AUM
 
 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with 5-PCF2Am-InsP5 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, Diphosphoinositol polyphosphate phosphohydrolase DDP1 | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
