8DYP
 
 | |
6FLP
 
 | |
6FLQ
 
 | |
6ZM9
 
 | | PHAGE SAM LYASE IN COMPLEX WITH S-METHYL-5'-THIOADENOSINE | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A, PHOSPHATE ION, ... | | Authors: | Guo, X, Kanchugal P, S, Selmer, M. | | Deposit date: | 2020-07-02 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a phage-encoded SAM lyase revises catalytic function of enzyme family.
Elife, 10, 2021
|
|
6ZMG
 
 | |
7YLQ
 
 | | Crystal structure of Microcystinase C from Sphingomonas sp. ACM-3962 at 2.6 A resolution | | Descriptor: | CALCIUM ION, Microcystinase C, ZINC ION | | Authors: | Guo, X, Li, Z, Ding, W, Yin, P, Feng, L. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Microcystin C from Sphingomonas sp. ACM-3962 at 2.6 A resolution
To Be Published
|
|
4U7T
 
 | | Crystal structure of DNMT3A-DNMT3L in complex with histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Guo, X, Wang, L, Yin, X, Li, J, Xiao, J, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
4ADO
 
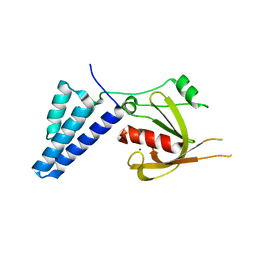 | | Fusidic acid resistance protein FusB | | Descriptor: | FAR1, ZINC ION | | Authors: | Guo, X, Peisker, K, Backbro, K, Chen, Y, Kiran, R.K, Sanyal, S, Selmer, M. | | Deposit date: | 2011-12-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of Fusb: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling
Open Biol., 2, 2012
|
|
4ADN
 
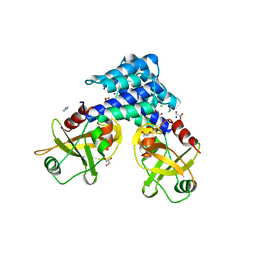 | | Fusidic acid resistance protein FusB | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FAR1, ... | | Authors: | Guo, X, Peisker, K, Backbro, K, Chen, Y, Kiran, R.K, Sanyal, S, Selmer, M. | | Deposit date: | 2011-12-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of Fusb: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling
Open Biol., 2, 2012
|
|
8KCP
 
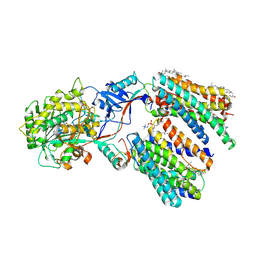 | | Cryo-EM structure of human gamma-secretase in complex with Crenigacestat | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of gamma-secretase inhibition by anti-cancer clinical drugs
To Be Published
|
|
8KCO
 
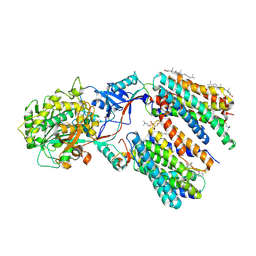 | | Cryo-EM structure of human gamma-secretase in complex with RO4929097 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,2-dimethyl-N-[(7S)-6-oxo-5,7-dihydrobenzo[d][1]benzazepin-7-yl]-N'-(2,2,3,3,3-pentafluoropropyl)propanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560
To Be Published
|
|
8KCU
 
 | | Cryo-EM structure of human gamma-secretase in complex with MK-0752 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural Basis of gamma-secretase inhibition by anti-cancer clinical drugs
To Be Published
|
|
8KCT
 
 | | Cryo-EM structure of human gamma-secretase in complex with Nirogacestat | | Descriptor: | (2S)-2-[[(2S)-6,8-bis(fluoranyl)-1,2,3,4-tetrahydronaphthalen-2-yl]amino]-N-[1-[1-(2,2-dimethylpropylamino)-2-methyl-propan-2-yl]imidazol-4-yl]pentanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of gamma-secretase inhibition by anti-cancer clinical drugs
To Be Published
|
|
8KCS
 
 | | Cryo-EM structure of human gamma-secretase in complex with BMS906024 | | Descriptor: | (2S,3R)-N'-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-2,3-bis[3,3,3-tris(fluoranyl)propyl]butanediamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural Basis of gamma-secretase inhibition by anti-cancer clinical drugs
To Be Published
|
|
8X54
 
 | | Cryo-EM structure of human gamma-secretase in complex with APP-C99 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
8X52
 
 | | Cryo-EM structure of human gamma-secretase in complex with Abeta49 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
8X53
 
 | | Cryo-EM structure of human gamma-secretase in complex with Abeta46 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
7Y5X
 
 | | CryoEM structure of PS2-containing gamma-secretase treated with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5T
 
 | | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5Z
 
 | | CryoEM structure of human PS2-containing gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
6IQD
 
 | | Crystal structure of Alcohol dehydrogenase from Geobacillus stearothermophilus | | Descriptor: | Alcohol dehydrogenase, ZINC ION | | Authors: | Xue, S, Feng, Y, Guo, X, Zhao, Z. | | Deposit date: | 2018-11-07 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Characterization of the substrate scope of an alcohol dehydrogenase commonly used as methanol dehydrogenase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5L6U
 
 | | S. ENTERICA HISA MUTANT - D10G, DUP13-15, Q24L, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Selmer, M. | | Deposit date: | 2016-05-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5L9F
 
 | | S. enterica HisA mutant - D10G, G11D, dup13-15, G44E, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SODIUM ION, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Selmer, M. | | Deposit date: | 2016-06-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4U7P
 
 | | Crystal structure of DNMT3A-DNMT3L complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, L, Guo, X, Li, J, Xiao, J, Yin, X, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.821 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
5ABT
 
 | | S.enterica HisA mutant D7N, G102A, V106M, D176A | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Protein Structures, Functions and Dynamics of Adaptive Evolution
To be Published
|
|
