3HLA
 
 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN A2.1 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A2.1) (ALPHA CHAIN) | | Authors: | Saper, M.A, Bjorkman, P.J, Wiley, D.C. | | Deposit date: | 1989-10-06 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution.
J.Mol.Biol., 219, 1991
|
|
8EDD
 
 | | Staphylococcus aureus endonuclease IV Y33F mutant | | Descriptor: | CHLORIDE ION, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Saper, M.A, Kirillov, S, Isupov, M.N, Wiener, R, Rouvinski, A. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Octahedral Iron in Catalytic Sites of Endonuclease IV from Staphylococcus aureus and Escherichia coli.
Biochemistry, 2024
|
|
8RLY
 
 | | E. coli endonuclease IV complexed with sulfate, catalytic Fe2+ | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Endonuclease 4, ... | | Authors: | Saper, M.A, Paterson, N.G, Kirillov, S, Rouvinski, A. | | Deposit date: | 2024-01-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Octahedral Iron in Catalytic Sites of Endonuclease IV from Staphylococcus aureus and Escherichia coli .
Biochemistry, 64, 2025
|
|
6U2A
 
 | |
3P42
 
 | |
5VAT
 
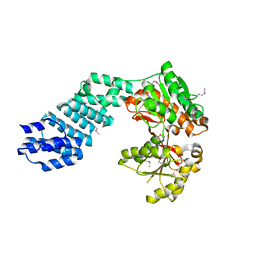 | |
1VHR
 
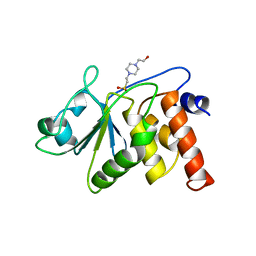 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE VHR, SULFATE ION | | Authors: | Yuvaniyama, J, Denu, J.M, Dixon, J.E, Saper, M.A. | | Deposit date: | 1996-02-20 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the dual specificity protein phosphatase VHR.
Science, 272, 1996
|
|
5KCN
 
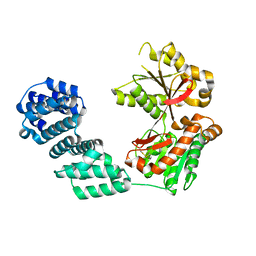 | |
1JLX
 
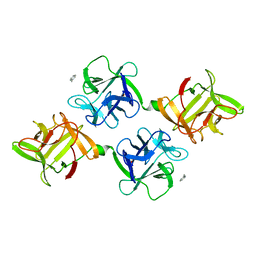 | | AGGLUTININ IN COMPLEX WITH T-DISACCHARIDE | | Descriptor: | AGGLUTININ, FORMYL GROUP, TOLUENE, ... | | Authors: | Transue, T.R, Smith, A.K, Mo, H, Goldstein, I.J, Saper, M.A. | | Deposit date: | 1997-07-23 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of benzyl T-antigen disaccharide bound to Amaranthus caudatus agglutinin.
Nat.Struct.Biol., 4, 1997
|
|
2HLA
 
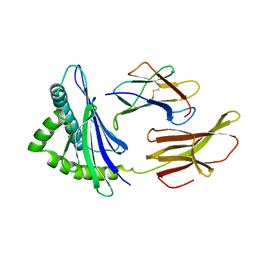 | |
1M0V
 
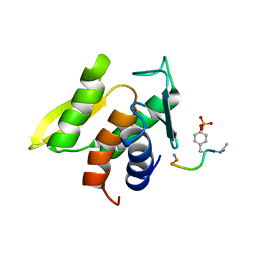 | | NMR STRUCTURE OF THE TYPE III SECRETORY DOMAIN OF YERSINIA YOPH COMPLEXED WITH THE SKAP-HOM PHOSPHO-PEPTIDE N-acetyl-DEpYDDPF-NH2 | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, SKAP55 homologue | | Authors: | Khandelwal, P, Keliikuli, K, Smith, C.L, Saper, M.A, Zuiderweg, E.R.P. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Biochemistry, 41, 2002
|
|
3CKM
 
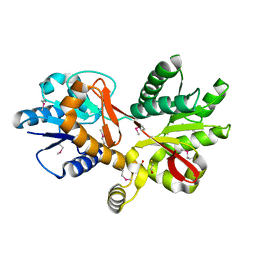 | |
4P29
 
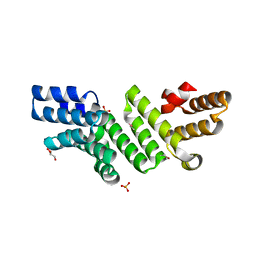 | |
5VBG
 
 | |
2I42
 
 | |
2LIV
 
 | |
6DR3
 
 | | Crystal structure of E. coli LpoA amino terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Kelley, A.C, Saper, M.A. | | Deposit date: | 2018-06-11 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structures of the amino-terminal domain of LpoA from Escherichia coli and Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6DCJ
 
 | |
1YTN
 
 | | HYDROLASE | | Descriptor: | NITRATE ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Yuvaniyama, C, Fauman, E.B, Saper, M.A. | | Deposit date: | 1996-05-01 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structures of Yersinia tyrosine phosphatase with bound tungstate and nitrate. Mechanistic implications.
J.Biol.Chem., 271, 1996
|
|
1YTW
 
 | | YERSINIA PTPASE COMPLEXED WITH TUNGSTATE | | Descriptor: | SULFATE ION, TUNGSTATE(VI)ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Fauman, E.B, Schubert, H.L, Saper, M.A. | | Deposit date: | 1996-05-01 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structures of Yersinia tyrosine phosphatase with bound tungstate and nitrate. Mechanistic implications.
J.Biol.Chem., 271, 1996
|
|
1YTS
 
 | | A LIGAND-INDUCED CONFORMATIONAL CHANGE IN THE YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Descriptor: | SULFATE ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Schubert, H.L, Stuckey, J.A, Fauman, E.B, Dixon, J.E, Saper, M.A. | | Deposit date: | 1995-04-07 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ligand-induced conformational change in the Yersinia protein tyrosine phosphatase.
Protein Sci., 4, 1995
|
|
8AXY
 
 | | Staphylococcus aureus endonuclease IV orthorhombic crystal form | | Descriptor: | FE (III) ION, Probable endonuclease 4, SULFATE ION, ... | | Authors: | Rouvinski, A, Kirillov, S, Saper, M.A, Wiener, R, Isupov, M.N. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Octahedral Iron in Catalytic Sites of Endonuclease IV from Staphylococcus aureus and Escherichia coli.
Biochemistry, 2024
|
|
1DM9
 
 | | HEAT SHOCK PROTEIN 15 KD | | Descriptor: | HYPOTHETICAL 15.5 KD PROTEIN IN MRCA-PCKA INTERGENIC REGION, SULFATE ION | | Authors: | Staker, B.L, Korber, P, Bardwell, J.C.A, Saper, M.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hsp15 reveals a novel RNA-binding motif.
EMBO J., 19, 2000
|
|
1EIZ
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE | | Descriptor: | FTSJ, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
1EJ0
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE, MERCURY DERIVATIVE | | Descriptor: | FTSJ, MERCURY (II) ION, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
