4ZWG
 
 | | Crystal structure of the GTP-dATP-bound catalytic core of SAMHD1 phosphomimetic T592E mutant | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tang, C, Ji, X, Xiong, Y. | | Deposit date: | 2015-05-19 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impaired dNTPase Activity of SAMHD1 by Phosphomimetic Mutation of Thr-592.
J.Biol.Chem., 290, 2015
|
|
4ZWE
 
 | |
1UPH
 
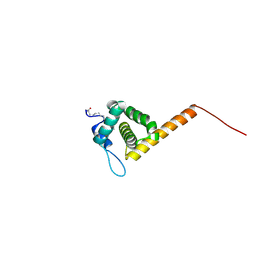 | | HIV-1 Myristoylated Matrix | | Descriptor: | GAG POLYPROTEIN | | Authors: | Tang, C, Loeliger, E, Luncsford, P, Kinde, I, Beckett, D, Summers, M.F. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-08 | | Last modified: | 2025-04-09 | | Method: | SOLUTION NMR | | Cite: | Entropic Switch Regulates Myristate Exposure in the HIV-1 Matrix Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1WCR
 
 | |
1GWP
 
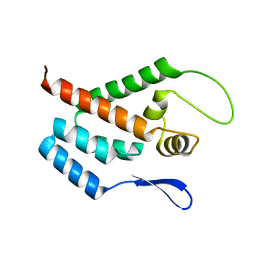 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE MATURE HIV-1 CAPSID PROTEIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Tang, C, Gitti, R.K, Lee, B.M, Walker, J, Summers, M.F, Yoo, S, Sundquist, W.I. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal 283-Residue Fragment of the Immature HIV-1 Gag Polyprotein
Nat.Struct.Biol., 9, 2002
|
|
5IW3
 
 | | anti-CD20 monoclonal antibody Fc fragment | | Descriptor: | ACETATE ION, Ig gamma-1 chain C region, SULFATE ION, ... | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of anti-CD20 monoclonal antibody Fc fragment at 2.05 Angstroms resolution
To Be Published
|
|
5IW6
 
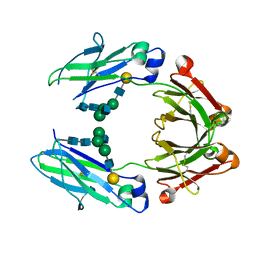 | | anti-CD20 monoclonal antibody Fc fragment | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2016-03-22 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of anti-CD20 monoclonal antibody Fc fragment at 2.34 Angstroms resolution
To Be Published
|
|
1L6N
 
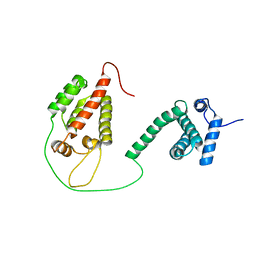 | |
2MZY
 
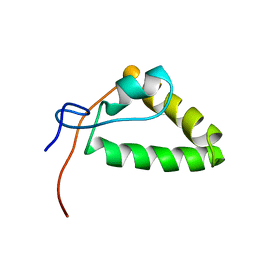 | | 1H, 13C, and 15N Chemical Shift Assignments and structure of Probable Fe(2+)-trafficking protein from Burkholderia pseudomallei 1710b. | | Descriptor: | Probable Fe(2+)-trafficking protein | | Authors: | Tang, C, Yang, F, Barnwal, R, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Probable Fe(2+)-trafficking protein
To be Published
|
|
2MRC
 
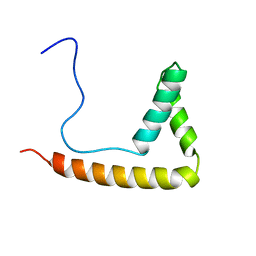 | |
6JDS
 
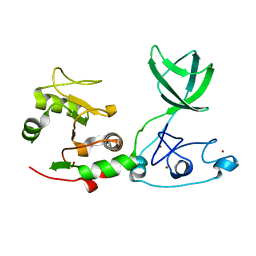 | |
6JDU
 
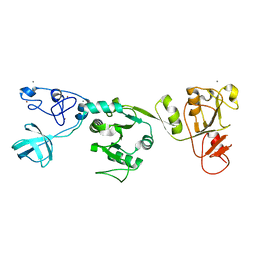 | | Crystal structure of PRRSV nsp10 (helicase) | | Descriptor: | CALCIUM ION, PP1b, ZINC ION | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2019-02-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
2V93
 
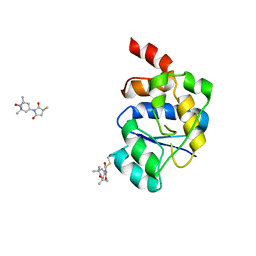 | | EQUILLIBRIUM MIXTURE OF OPEN AND PARTIALLY-CLOSED SPECIES IN THE APO STATE OF MALTODEXTRIN-BINDING PROTEIN BY PARAMAGNETIC RELAXATION ENHANCEMENT NMR | | Descriptor: | 1-(1-HYDROXY-2,2,6,6-TETRAMETHYLPIPERIDIN-4-YL)PYRROLIDINE-2,5-DIONE, MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Clore, G.M, Tang, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR.
Nature, 449, 2007
|
|
6JDR
 
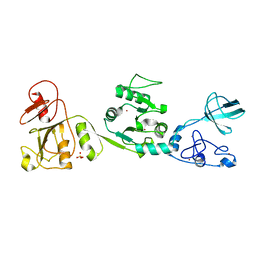 | |
6WEM
 
 | | Crimson 0.9 | | Descriptor: | mCrimson 0.9 | | Authors: | Ataie, N, Tran Tang, C, Sens, A, Lin, M.Z, Chu, J, Ng, H.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crimson 0.9
To Be Published
|
|
4BQ8
 
 | | Crystal structure of the RGMB-NEO1 complex form 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
4BQC
 
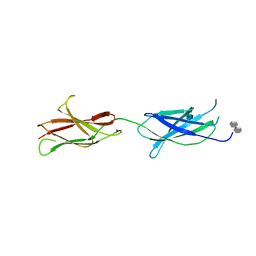 | | Crystal structure of the FN5 and FN6 domains of NEO1 bound to SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, ... | | Authors: | Bell, C.H, Healey, E, vanErp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
4BQ7
 
 | | Crystal structure of the RGMB-Neo1 complex form 2 | | Descriptor: | NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (6.601 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
5YMY
 
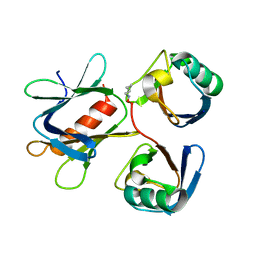 | | The structure of the complex between Rpn13 and K48-diUb | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Liu, Z, Dong, X, Gong, Z, Yi, H.W, Liu, K, Yang, J, Zhang, W.P, Tang, C. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13.
Cell Discov, 5, 2019
|
|
5YZ9
 
 | | zinc finger domain of METTL3-METTL14 N6-methyladenosine methyltransferase | | Descriptor: | N6-adenosine-methyltransferase catalytic subunit, ZINC ION | | Authors: | Dong, X, Tang, C, Gong, Z, Yin, P, Huang, J.B. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase.
Protein Cell, 10, 2019
|
|
1EII
 
 | | NMR STRUCTURE OF HOLO CELLULAR RETINOL-BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II, RETINOL | | Authors: | Lu, J, Lin, C.L, Tang, C, Ponder, J.W, Kao, J.L, Cistola, D.P, Li, E. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding of retinol induces changes in rat cellular retinol-binding protein II conformation and backbone dynamics.
J.Mol.Biol., 300, 2000
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1VRV
 
 | |
4BQ6
 
 | | Crystal structure of the RGMB-NEO1 complex form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
