2DFX
 
 | | Crystal structure of the carboxy terminal domain of colicin E5 complexed with its inhibitor | | Descriptor: | Colicin-E5, Colicin-E5 immunity protein | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-03-06 | | Release date: | 2007-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
2DJH
 
 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-04-03 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
1T1S
 
 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
1TFK
 
 | | Ribonuclease from Escherichia coli complexed with its inhibtor protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
1TFO
 
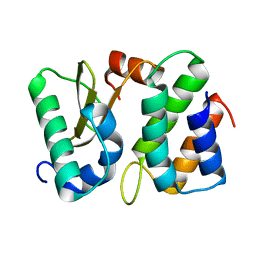 | | Ribonuclease from Escherichia coli complexed with its inhibitor protein | | Descriptor: | Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
1T1R
 
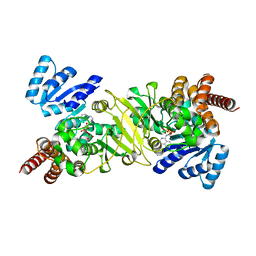 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, SULFATE ION, [(ISOQUINOLIN-1-YLAMINO)-PHOSPHONO-METHYL]-PHOSPHONIC ACID | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
1JVS
 
 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase; a target enzyme for antimalarial drugs | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yajima, S, Nonaka, T, Kuzuyama, T, Seto, H, Ohsawa, K. | | Deposit date: | 2001-08-31 | | Release date: | 2002-10-09 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with cofactors: implications of a flexible loop movement upon substrate binding.
J.Biochem., 131, 2002
|
|
2EGH
 
 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with a magnesium ion, NADPH and fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | Authors: | Yajima, S, Hara, K, Iino, D, Sasaki, Y, Kuzuyama, T, Seto, H. | | Deposit date: | 2007-03-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in a quaternary complex with a magnesium ion, NADPH and the antimalarial drug fosmidomycin
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3VJ7
 
 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 R33Q mutant | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Fushinobu, S, Ogawa, T, Hidaka, M, Masaki, H. | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the catalytic residues of sequence-specific and histidine-free ribonuclease colicin E5
J.Biochem., 152, 2012
|
|
2JLN
 
 | | Structure of Mhp1, a nucleobase-cation-symport-1 family transporter | | Descriptor: | MERCURY (II) ION, MHP1, SODIUM ION | | Authors: | Weyand, S, Shimamura, T, Yajima, S, Suzuki, S, Mirza, O, Krusong, K, Carpenter, E.P, Rutherford, N.G, Hadden, J.M, O'Reilly, J, Ma, P, Saidijam, M, Patching, S.G, Hope, R.J, Norbertczak, H.T, Roach, P.C.J, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2008-09-11 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Molecular Mechanism of a Nucleobase-Cation-Symport-1 Family Transporter.
Science, 322, 2008
|
|
5B2H
 
 | | Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi | | Descriptor: | HA-33, TRIETHYLENE GLYCOL | | Authors: | Akiyama, T, Hayashi, S, Matsumoto, T, Hasegawa, K, Yamano, A, Suzuki, T, Niwa, K, Watanabe, T, Sagane, Y, Yajima, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex
Biochem.Biophys.Res.Commun., 476, 2016
|
|
3WG8
 
 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist AS6 | | Descriptor: | (2Z,4E)-5-[(1S)-3-(hexylsulfanyl)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Okamoto, M, Muto, T, Endo, A, Nambara, E, Hirai, N, Ohnishi, T, Cutler, S.R, Todoroki, Y, Yajima, S. | | Deposit date: | 2013-07-31 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions
Nat.Chem.Biol., 10, 2014
|
|
7CUO
 
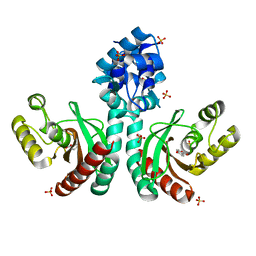 | | IclR transcription factor complexed with 4-hydroxybenzoic acid from Microbacterium hydrocarbonoxydans | | Descriptor: | P-HYDROXYBENZOIC ACID, SULFATE ION, Transcription factor | | Authors: | Akiyama, T, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-08-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
7D5M
 
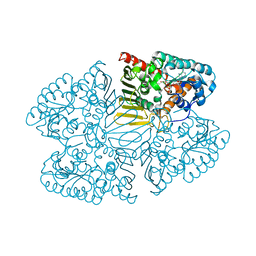 | | Crystal structure of inositol dehydrogenase homolog complexed with NAD+ from Azotobacter vinelandii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NAD+ from Azotobacter vinelandii
To Be Published
|
|
7D5N
 
 | | Crystal structure of inositol dehydrogenase homolog complexed with NADH and myo-inositol from Azotobacter vinelandii | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NADH and myo-inositol from Azotobacter vinelandii
To Be Published
|
|
7DQB
 
 | | Crystal structure of an IclR homolog complexed with 4-hydroxybenzoate from Microbacterium hydrocarbonoxydans in P212121 form | | Descriptor: | IclR homolog, P-HYDROXYBENZOIC ACID | | Authors: | Akiyama, T, Sasaki, Y, Ito, S, Yajima, S. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
2DGA
 
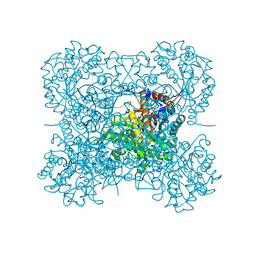 | | Crystal structure of hexameric beta-glucosidase in wheat | | Descriptor: | Beta-glucosidase, GLYCEROL, SULFATE ION | | Authors: | Sue, M, Yamazaki, K, Miyamoto, T, Yajima, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and Structural Characterization of Hexameric beta-D-Glucosidases in Wheat and Rye.
Plant Physiol., 141, 2006
|
|
5H1A
 
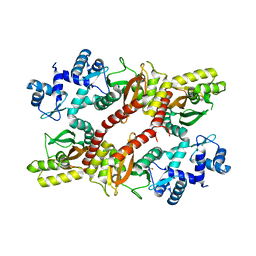 | | Crystal structure of an IclR homolog from Microbacterium sp. strain HM58-2 | | Descriptor: | IclR transcription factor homolog, PHOSPHATE ION | | Authors: | Akiyama, T, Yamada, Y, Takaya, N, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an IclR homologue from Microbacterium sp. strain HM58-2.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6IY9
 
 | | Crystal structure of aminoglycoside 7"-phoshotransferase-Ia (APH(7")-Ia/HYG) from Streptomyces hygroscopicus complexed with hygromycin B | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, HYGROMYCIN B VARIANT, ... | | Authors: | Takenoya, M, Shimamura, T, Yamanaka, R, Adachi, Y, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2018-12-14 | | Release date: | 2019-09-11 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the substrate recognition of aminoglycoside 7''-phosphotransferase-Ia from Streptomyces hygroscopicus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6KTK
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTL
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTJ
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, apo-form, from Paracoccus laeviglucosivorans | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6AA8
 
 | |
6ACQ
 
 | |
6LU2
 
 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans | | Descriptor: | Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
