6P9B
 
 | |
6P9A
 
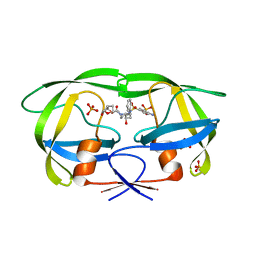 | | HIV-1 Protease multiple mutant PRS5B with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease, PHOSPHATE ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.
Febs J., 287, 2020
|
|
6PJG
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-97 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-alanyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PJC
 
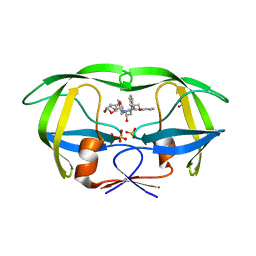 | | HIV-1 Protease NL4-3 WT in Complex with LR4-41 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-5-{[(2,6-dimethylphenoxy)acetyl]amino}-4-hydroxy-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.965 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PU8
 
 | | Room temperature X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate of keto-darunavir | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3,3-dihydroxy-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
1PRO
 
 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-98881 | | Descriptor: | (5R,6R)-2,4-BIS-(4-HYDROXY-3-METHOXYBENZYL)-1,5-DIBENZYL-3-OXO-6-HYDROXY-1,2,4-TRIAZACYCLOHEPTANE, HIV-1 PROTEASE | | Authors: | Park, C.H, Kong, X.P, Dealwis, C.G. | | Deposit date: | 1995-07-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel, picomolar inhibitor of human immunodeficiency virus type 1 protease.
J.Med.Chem., 39, 1996
|
|
1QBT
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6ALPHA,7ALPHA)]-3,3'-{{TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-DIYL]BIS(METHYLENE)]BIS[N-1H-BENZIMIDAZOL-2-YLBENZAMIDE] | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
6PTP
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | HIV-1 Protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
1QBR
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
1QBS
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic HIV protease inhibitors: synthesis, conformational analysis, P2/P2' structure-activity relationship, and molecular recognition of cyclic ureas.
J.Med.Chem., 39, 1996
|
|
1Q9P
 
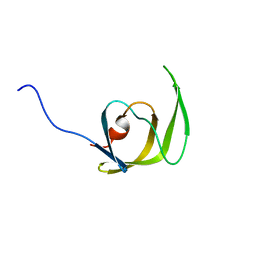 | | Solution structure of the mature HIV-1 protease monomer | | Descriptor: | HIV-1 Protease | | Authors: | Ishima, R, Torchia, D.A, Lynch, S.M, Gronenborn, A.M, Louis, J.M. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mature HIV-1 protease monomer: Insight into the tertiary fold and stability of a precursor
J.Biol.Chem., 278, 2003
|
|
1QBU
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R--(1ALPHA,5ALPHA,7BETA)]-3-[(CYCLOPROPHYLMETHYL)HEXAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPIN] METHYL-2-THIAZOLYLBENZAMIDE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
1RV7
 
 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-12 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1RPI
 
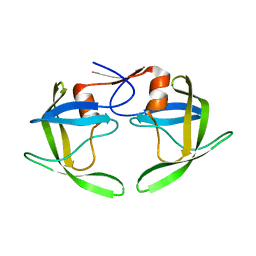 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | alpha-D-glucopyranose, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-03 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1RL8
 
 | | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir | | Descriptor: | RITONAVIR, protease RETROPEPSIN | | Authors: | Rezacova, P, Brynda, J, Sedlacek, J, Konvalinka, J, Fabry, M, Horejsi, M. | | Deposit date: | 2003-11-25 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir
To be Published, 2005
|
|
1RQ9
 
 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-04 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1SBG
 
 | |
1SDV
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
6S1U
 
 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
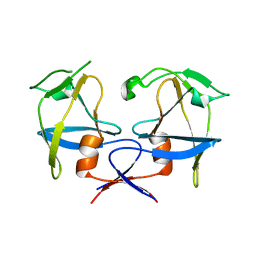 | | Crystal structure of dimeric M-PMV protease D26N mutant | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1SP5
 
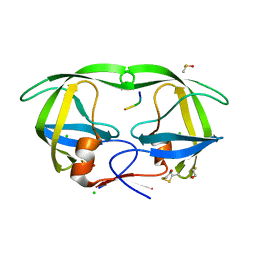 | | Crystal structure of HIV-1 protease complexed with a product of autoproteolysis | | Descriptor: | 5-mer peptide from Protease, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Vondrackova, E, Hasek, J, Jaskolski, M, Rezacova, P, Dohnalek, J, Skalova, T, Petrokova, H, Duskova, J, Brynda, J, Sedlacek, J. | | Deposit date: | 2004-03-16 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Product of enzymatic self-cleavage bound in the active site of HIV protease
To be Published
|
|
1SGU
 
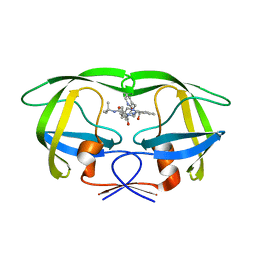 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, Mckenna, R, Abandje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
1SDT
 
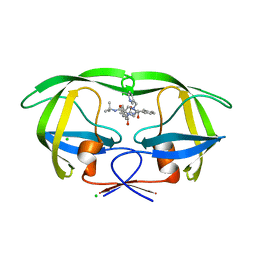 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
1SIV
 
 | |
1SDU
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | ACETATE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, SULFATE ION, ... | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
