4OQA
 
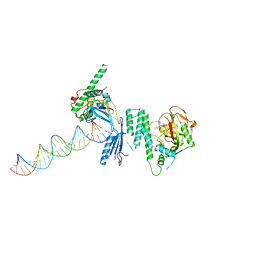 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2Z)-2-(2,4-dihydroxybenzylidene)-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide | | Descriptor: | (2Z)-2-(2,4-dihydroxybenzylidene)-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Steffen, J.D. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
2JVN
 
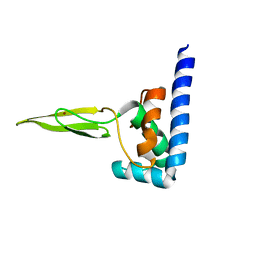 | | Domain C of human PARP-1 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Hoffman, D, Tao, Z, Liu, H. | | Deposit date: | 2007-09-24 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif.
Biochemistry, 47, 2008
|
|
4XHU
 
 | |
6WZ9
 
 | |
3L3M
 
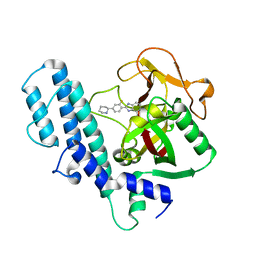 | | PARP complexed with A927929 | | Descriptor: | 2-{2-fluoro-4-[(2S)-piperidin-2-yl]phenyl}-1H-benzimidazole-7-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of phenyl-substituted benzimidazole carboxamide poly(ADP-ribose) polymerase inhibitors: identification of (S)-2-(2-fluoro-4-(pyrrolidin-2-yl)phenyl)-1H-benzimidazole-4-carboxamide (A-966492), a highly potent and efficacious inhibitor.
J.Med.Chem., 53, 2010
|
|
3L3L
 
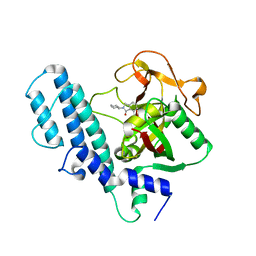 | | PARP complexed with A906894 | | Descriptor: | 3-oxo-2-piperidin-4-yl-2,3-dihydro-1H-isoindole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and SAR of substituted 3-oxoisoindoline-4-carboxamides as potent inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6VKQ
 
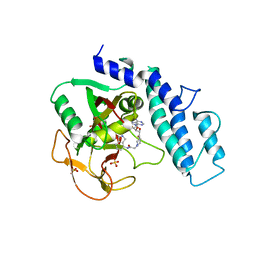 | | Crystal Structure of human PARP-1 CAT domain bound to inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Steffen, J.D, Pascal, J.M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for allosteric PARP-1 retention on DNA breaks.
Science, 368, 2020
|
|
6VKK
 
 | |
6VKO
 
 | |
6TX1
 
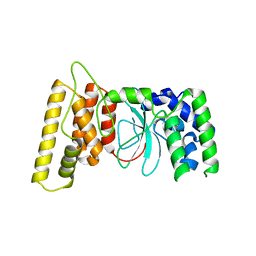 | | HPF1 from Nematostella vectensis | | Descriptor: | Predicted protein | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
6TVH
 
 | |
6TX2
 
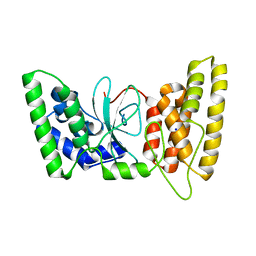 | | Human HPF1 | | Descriptor: | Histone PARylation factor 1, SODIUM ION | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
6I8T
 
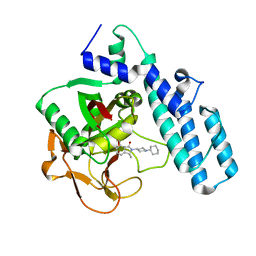 | |
6I8M
 
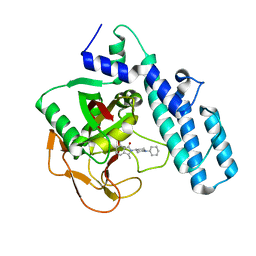 | |
2RCW
 
 | | PARP complexed with A620223 | | Descriptor: | Poly [ADP-ribose] polymerase 1, trans-4-(7-carbamoyl-1H-benzimidazol-2-yl)-1-propylpiperidinium | | Authors: | Park, C.H. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | PARP complexed with A620223
To be Published
|
|
1EFY
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC FRAGMENT OF POLY (ADP-RIBOSE) POLYMERASE COMPLEXED WITH A BENZIMIDAZOLE INHIBITOR | | Descriptor: | 2-(3'-METHOXYPHENYL) BENZIMIDAZOLE-4-CARBOXAMIDE, POLY (ADP-RIBOSE) POLYMERASE | | Authors: | White, A.W, Almassy, R, Calvert, A.H, Curtin, N.J, Griffin, R.J, Hostomsky, Z, Maegley, K, Newell, D.R, Srinivasan, S, Golding, B.T. | | Deposit date: | 2000-02-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase.
J.Med.Chem., 43, 2000
|
|
2RD6
 
 | | PARP complexed with A861695 | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2007-09-21 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PARP complexed with A861695
To be Published
|
|
2RIQ
 
 | | Crystal Structure of the Third Zinc-binding domain of human PARP-1 | | Descriptor: | ETHANOL, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.F, Servent, K.M. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Third Zinc-binding Domain of Human Poly(ADP-ribose) Polymerase-1 Coordinates DNA-dependent Enzyme Activation.
J.Biol.Chem., 283, 2008
|
|
1PAX
 
 | |
6NRI
 
 | | Crystal Structure of human PARP-1 ART domain bound to inhibitor UTT83 | | Descriptor: | (2Z)-2-{[4-(3-cyclopropyl-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazine-7(8H)-carbonyl)phenyl]methylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
5DS3
 
 | | Crystal structure of constitutively active PARP-1 | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, PENTAETHYLENE GLYCOL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
2CR9
 
 | |
2COK
 
 | |
2CS2
 
 | |
3OD8
 
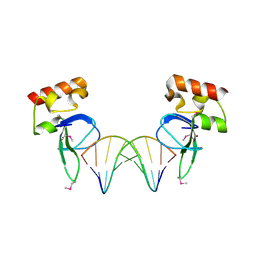 | | Human PARP-1 zinc finger 1 (Zn1) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*AP*GP*CP*GP*GP*C)-3', 5'-D(*GP*CP*CP*GP*CP*TP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
