5I4U
 
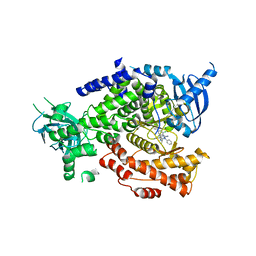 | | The crystal structure of PI3Kdelta with compound 34 | | Descriptor: | 2,4-diamino-6-{[(1S)-1-(5-chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)ethyl]amino}pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A.G. | | Deposit date: | 2016-02-12 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | The Design and Synthesis of Potent, Selective and Metabolically Stable PI3K[delta] Inhibitors
To be published
|
|
5I6U
 
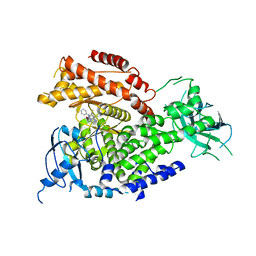 | | The crystal structure of PI3Kdelta with compound 32 | | Descriptor: | 2-[(1S)-1-({6-amino-5-[(1H-pyrazol-4-yl)ethynyl]pyrimidin-4-yl}amino)ethyl]-5-chloro-3-phenylquinazolin-4(3H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A.G. | | Deposit date: | 2016-02-16 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | The crystal structure of PI3Kdelta with compound 32
To Be Published
|
|
5IS5
 
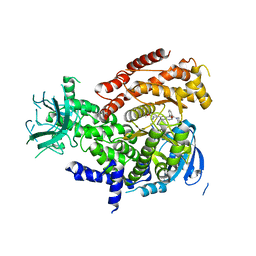 | |
5ITD
 
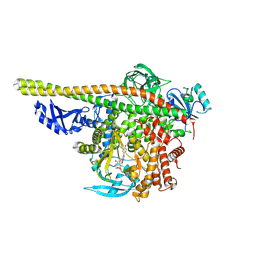 | | Crystal structure of PI3K alpha with PI3K delta inhibitor | | Descriptor: | 5-{4-[3-(4-acetylpiperazine-1-carbonyl)phenyl]quinazolin-6-yl}-2-methoxypyridine-3-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Discovery and Pharmacological Characterization of Novel Quinazoline-Based PI3K Delta-Selective Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5JHA
 
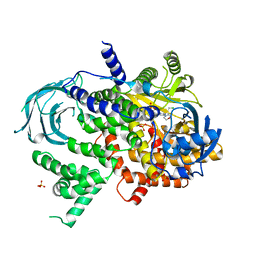 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin2 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)pyrimidin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5JHB
 
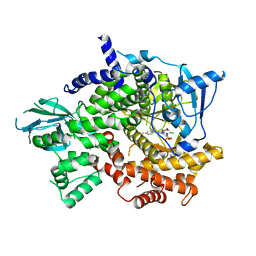 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5KAE
 
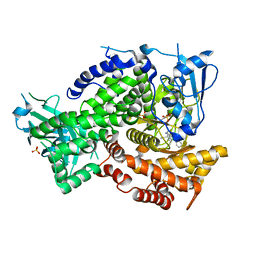 | |
5KC2
 
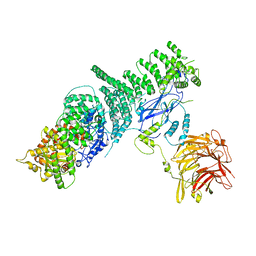 | | Negative stain structure of Vps15/Vps34 complex | | Descriptor: | Phosphatidylinositol 3-kinase VPS34, Serine/threonine-protein kinase VPS15 | | Authors: | Kirsten, M.L, Zhang, L, Ohashi, Y, Perisic, O, Williams, R.L, Sachse, C. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
5L72
 
 | |
5LUQ
 
 | | Crystal Structure of Human DNA-dependent Protein Kinase Catalytic Subunit (DNA-PKcs) | | Descriptor: | C-terminal fragment of KU80 (KU80ct194), DNA-dependent protein kinase catalytic subunit,DNA-dependent Protein Kinase Catalytic Subunit,DNA-dependent protein kinase catalytic subunit | | Authors: | Sibanda, B.L, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-15 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair.
Science, 355, 2017
|
|
5M6U
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH LASW1579 | | Descriptor: | 4-azanyl-6-[[(1~{S})-1-(4-oxidanylidene-3-phenyl-pyrrolo[2,1-f][1,2,4]triazin-2-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Segarra, V, Hernandez, B, Lozoya, E, Blaesse, M, Hoeppner, S, Jestel, A. | | Deposit date: | 2016-10-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Available PI3K delta Inhibitor for the Treatment of Inflammatory Diseases.
ACS Med Chem Lett, 8, 2017
|
|
5NCY
 
 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-(3-methyl-5-oxidanylidene-6-phenyl-[1,3]thiazolo[3,2-a]pyridin-7-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
5NCZ
 
 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-[6-[3-[(dimethylamino)methyl]phenyl]-3-methyl-5-oxidanylidene-[1,3]thiazolo[3,2-a]pyridin-7-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
5NGB
 
 | |
5NP0
 
 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5NP1
 
 | | Open protomer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5O83
 
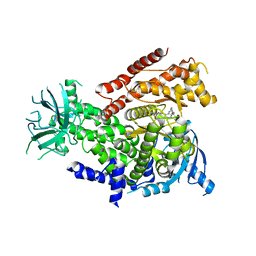 | | Discovery of CDZ173 (leniolisib), Representing a Structurally Novel Class of PI3K Delta-Selective Inhibitors | | Descriptor: | Leniolisib, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of CDZ173 (Leniolisib), Representing a Structurally Novel Class of PI3K Delta-Selective Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5OEJ
 
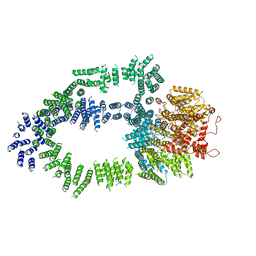 | | Structure of Tra1 subunit within the chromatin modifying complex SAGA | | Descriptor: | Tra1 subunit within the chromatin modifying complex SAGA | | Authors: | Sharov, G, Voltz, K, Durand, A, Kolesnikova, O, Papai, G, Myasnikov, A.G, Dejaegere, A, Ben-Shem, A, Schultz, P. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA.
Nat Commun, 8, 2017
|
|
5OJS
 
 | |
5OQ4
 
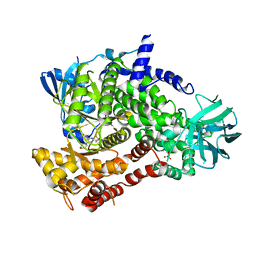 | | PQR309 - a Potent, Brain-Penetrant, Orally Bioavailable, pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology | | Descriptor: | 5-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Williams, R.L, Zhang, X. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology.
J. Med. Chem., 60, 2017
|
|
5SW8
 
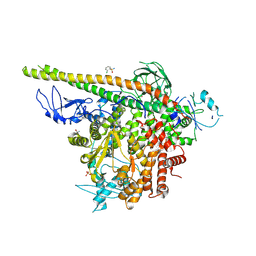 | | Crystal structure of PI3Kalpha in complex with fragments 7 and 11 | | Descriptor: | 2-CHLOROBENZENESULFONAMIDE, 2H-indazol-5-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SWG
 
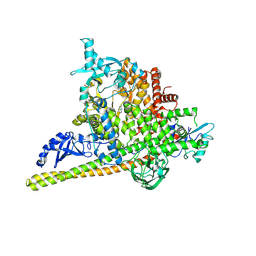 | | Crystal Structure of PI3Kalpha in complex with fragments 5 and 21 | | Descriptor: | 1H-benzimidazol-2-amine, CATECHOL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SWO
 
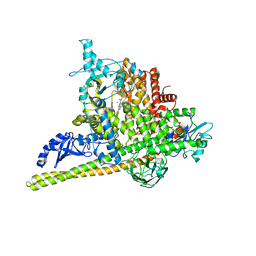 | | Crystal Structure of PI3Kalpha in complex with fragments 4 and 19 | | Descriptor: | 2-methyl-5-nitro-1H-indole, 4-methyl-3-nitropyridin-2-amine, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SWP
 
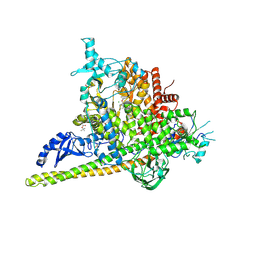 | | Crystal Structure of PI3Kalpha in complex with fragments 6 and 24 | | Descriptor: | 2-methylcyclohexane-1,3-dione, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SWR
 
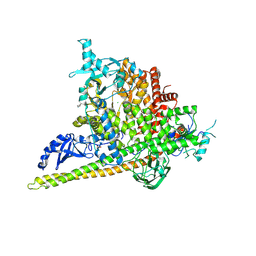 | | Crystal Structure of PI3Kalpha in complex with fragments 20 and 26 | | Descriptor: | 2-HYDROXYBENZOIC ACID, 6-hydroxy-3,4-dihydronaphthalen-1(2H)-one, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
