2F4J
 
 | | Structure of the Kinase Domain of an Imatinib-Resistant Abl Mutant in Complex with the Aurora Kinase Inhibitor VX-680 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Young, M.A, Shah, N.P, Chao, L.H, Zarrinkar, P, Sawyers, P, Kuriyan, J. | | Deposit date: | 2005-11-23 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680.
Cancer Res., 66, 2006
|
|
1SL3
 
 | | crystal structue of Thrombin in complex with a potent P1 heterocycle-Aryl based inhibitor | | Descriptor: | (2-[6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDOPYRIDIN-2-YL)ETHYL]AMINO}-2-OXOPYRAZIN-1(2H)-YL]-N-[5-CHLORO-2-(1H-TETRAZOL-1-YL)BENZYL]ACETAMIDE, Hirudin, thrombin | | Authors: | Young, M.B, Barrow, J.C, Glass, K.L, Lundell, G.F, Newton, C.L, Pellicore, J.M, Rittle, K.E, Selnick, H.G, Stauffer, K.J, Vacca, J.P, Williams, P.D, Bohn, D, Clayton, F.C, Cook, J.J, Krueger, J.A, Kuo, L.C, Lewis, S.D, Lucas, B.J, McMasters, D.R, Miller-Stein, C, Pietrak, B.L. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and evaluation of potent P1 aryl heterocycle-based thrombin inhibitors
J.Med.Chem., 47, 2004
|
|
3QHW
 
 | | Structure of a pCDK2/CyclinA transition-state mimic | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CDK2 substrate peptide: PKTPKKAKKL, ... | | Authors: | Young, M.A, Jacobsen, D.M, Bao, Z.Q. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Briefly Bound to Activate: Transient Binding of a Second Catalytic Magnesium Activates the Structure and Dynamics of CDK2 Kinase for Catalysis.
Structure, 19, 2011
|
|
3QHR
 
 | | Structure of a pCDK2/CyclinA transition-state mimic | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CDK2 substrate peptide: PKTPKKAKKL, CHLORIDE ION, ... | | Authors: | Young, M.A, Jacobsen, D.M, Bao, Z.Q. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Briefly Bound to Activate: Transient Binding of a Second Catalytic Magnesium Activates the Structure and Dynamics of CDK2 Kinase for Catalysis.
Structure, 19, 2011
|
|
4I3Z
 
 | | Structure of pCDK2/CyclinA bound to ADP and 2 Magnesium ions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Cyclin-A2, ... | | Authors: | Jacobsen, D.M, Bao, Z.-Q, O'Brien, P.J, Brooks, C.L, Young, M.A. | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Price To Be Paid for Two-Metal Catalysis: Magnesium Ions That Accelerate Chemistry Unavoidably Limit Product Release from a Protein Kinase
J.Am.Chem.Soc., 134, 2012
|
|
4II5
 
 | | Structure of PCDK2/CYCLINA bound to ADP and 1 MAGNESIUM ION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Jacobsen, D.M, Bao, Z.-Q, O'Brien, P.J, Brooks III, C.L, Young, M.A. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Price to be paid for two-metal catalysis: Magnesium ions that accelerate chemistry unavoidably limit product release from a PROTEIN KINASE
J.Am.Chem.Soc., 134, 2012
|
|
4GU5
 
 | | Structure of Full-length Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zoltowski, B.D, Vaidya, A.T, Top, D, Widom, J, Young, M.W, Levy, C, Jones, A.R, Scrutton, N.S, Leys, D, Crane, B.R. | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Updated structure of Drosophila cryptochrome.
Nature, 495, 2013
|
|
1ZA7
 
 | | The crystal structure of salt stable cowpea cholorotic mottle virus at 2.7 angstroms resolution. | | Descriptor: | Coat protein | | Authors: | Bothner, B, Speir, J.A, Qu, C, Willits, D.A, Young, M.J, Johnson, J.E. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics.
J.Virol., 80, 2006
|
|
2C0N
 
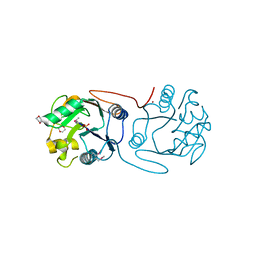 | | Crystal Structure of A197 from STIV | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, A197, NICKEL (II) ION, ... | | Authors: | Larson, E.T, Reiter, D, Young, M, Lawrence, C.M. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-29 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of A197 from Sulfolobus Turreted Icosahedral Virus: A Crenarchaeal Viral Glycosyltransferase Exhibiting the Gt-A Fold.
J.Virol., 80, 2006
|
|
6GYN
 
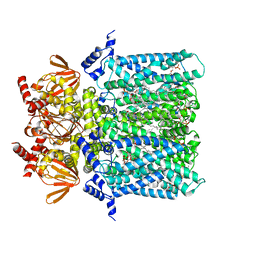 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
6GYO
 
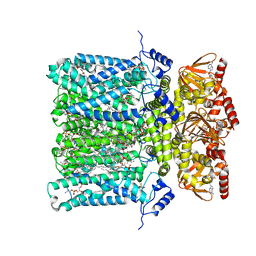 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel in complex with cAMP | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
3J31
 
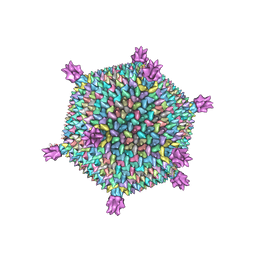 | | Life in the extremes: atomic structure of Sulfolobus Turreted Icosahedral Virus | | Descriptor: | A223 penton base, A55 membrane protein, C381 turret protein, ... | | Authors: | Veesler, D, Ng, T.S, Sendamarai, A.K, Eilers, B.J, Lawrence, C.M, Lok, S.M, Young, M.J, Johnson, J.E, Fu, C.-Y. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2CLB
 
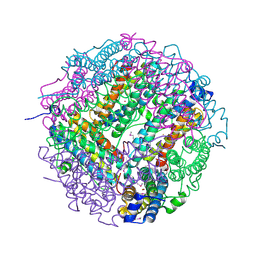 | | The structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like di-metal binding site within a Dps- like dodecameric assembly | | Descriptor: | DPS-LIKE PROTEIN, FE (III) ION, ZINC ION | | Authors: | Gauss, G.H, Benas, P, Wiedenheft, B, Young, M, Douglas, T, Lawrence, C.M. | | Deposit date: | 2006-04-26 | | Release date: | 2006-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Dps-Like Protein from Sulfolobus Solfataricus Reveals a Bacterioferritin-Like Dimetal Binding Site within a Dps-Like Dodecameric Assembly.
Biochemistry, 45, 2006
|
|
3UXU
 
 | |
2VZB
 
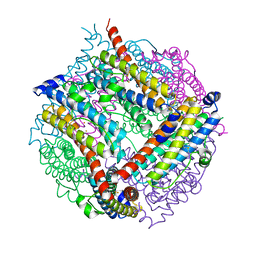 | | A Dodecameric Thioferritin in the Bacterial Domain, Characterization of the Bacterioferritin-Related Protein from Bacteroides fragilis | | Descriptor: | BENZAMIDINE, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Gauss, G.H, Young, M.J, Douglas, T, Lawrence, C.M. | | Deposit date: | 2008-07-31 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Bacteroides Fragilis Bfr Gene Product Identifies a Bacterial Dps-Like Protein and Suggests Evolutionary Links in the Ferritin Superfamily.
J.Bacteriol., 194, 2012
|
|
8SBK
 
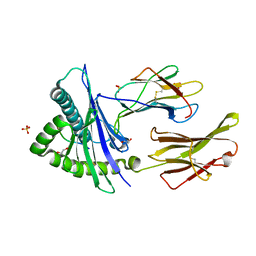 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI (ATG2A). | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, ... | | Authors: | Mallik, L, Young, M.C, Sgourakis, N.G. | | Deposit date: | 2023-04-03 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
2WBT
 
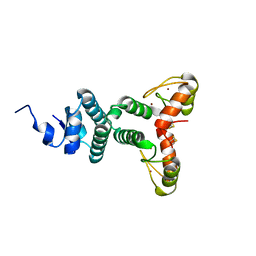 | | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA | | Descriptor: | B-129, ZINC ION | | Authors: | Eilers, B.J, Menon, S, Windham, A.B, Kraft, P, Dlakic, M, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA
To be Published
|
|
8SBL
 
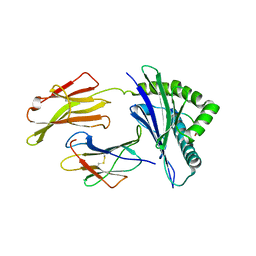 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI | | Descriptor: | Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, MHC class I antigen | | Authors: | Mallik, L, Young, M.C, Sgourakis, N.G. | | Deposit date: | 2023-04-03 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
1JQL
 
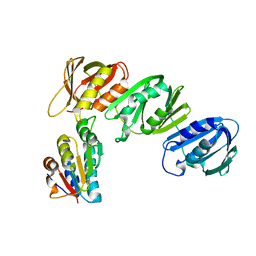 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of beta-delta (1-140) | | Descriptor: | DNA Polymerase III, BETA CHAIN, DELTA SUBUNIT | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
2W8M
 
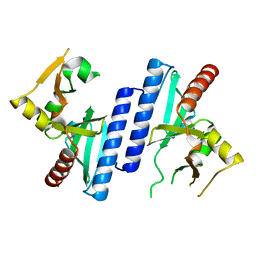 | |
1JQJ
 
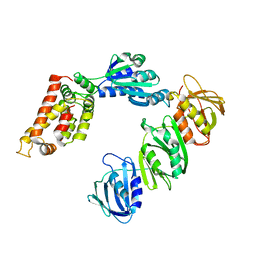 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of the beta-delta complex | | Descriptor: | DNA polymerase III, beta chain, delta subunit | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1TBX
 
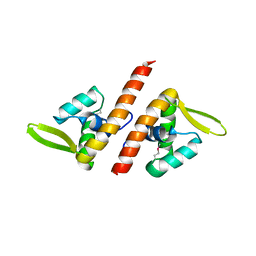 | | Crystal structure of SSV1 F-93 | | Descriptor: | Hypothetical 11.0 kDa protein | | Authors: | Kraft, P, Oeckinghaus, A, Kummel, D, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-05-20 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein.
J.Virol., 78, 2004
|
|
2WTE
 
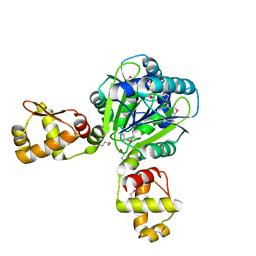 | | The structure of the CRISPR-associated protein, Csa3, from Sulfolobus solfataricus at 1.8 angstrom resolution. | | Descriptor: | CSA3, DI(HYDROXYETHYL)ETHER | | Authors: | Lintner, N.G, Alsbury, D.L, Copie, V, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Crispr-Associated Protein Csa3 Provides Insight Into the Regulation of the Crispr/Cas System.
J.Mol.Biol., 405, 2011
|
|
2VQC
 
 | | Structure of a DNA binding winged-helix protein, F-112, from Sulfolobus Spindle-shaped Virus 1. | | Descriptor: | HYPOTHETICAL 13.2 KDA PROTEIN | | Authors: | Menon, S.K, Kraft, P, Corn, G.J, Wiedenheft, B, Young, M.J, Lawrence, C.M. | | Deposit date: | 2008-03-12 | | Release date: | 2008-05-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Usage in Sulfolobus Spindle-Shaped Virus 1 and Extension to Hyperthermophilic Viruses in General.
Virology, 376, 2008
|
|
1SKV
 
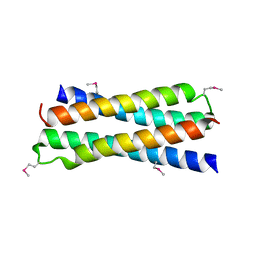 | | Crystal Structure of D-63 from Sulfolobus Spindle Virus 1 | | Descriptor: | Hypothetical 7.5 kDa protein | | Authors: | Kraft, P, Kummel, D, Oeckinghaus, A, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of d-63 from sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function
J.Virol., 78, 2004
|
|
