5V4V
 
 | | Saccharomyces cerevisiae Old Yellow Enzyme 3 | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, NADPH dehydrogenase 3, ... | | Authors: | Stewart, J.D. | | Deposit date: | 2017-03-10 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Saccharomyces cerevisiae Old Yellow Enzyme 3
To Be Published
|
|
5V4P
 
 | |
1BDK
 
 | | AN NMR, CD, MOLECULAR DYNAMICS, AND FLUOROMETRIC STUDY OF THE CONFORMATION OF THE BRADYKININ ANTAGONIST B-9340 IN WATER AND IN AQUEOUS MICELLAR SOLUTIONS | | Descriptor: | bradykinin antagonist B-9340 | | Authors: | Sejbal, J, Kotovych, G, Cann, J.R, Stewart, J.M, Gera, L. | | Deposit date: | 1995-07-28 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | An NMR, CD, molecular dynamics, and fluorometric study of the conformation of the bradykinin antagonist B-9340 in water and in aqueous micellar solutions.
J.Med.Chem., 39, 1996
|
|
4QAI
 
 | | P. stipitis OYE2.6-Y78W | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase | | Authors: | Sullivan, B, Walton, A.Z, Stewart, J.D. | | Deposit date: | 2014-05-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Residues Controlling Facial Selectivity in an Alkene Reductase and Semirational Alterations to Create Stereocomplementary Variants.
ACS CATALYSIS, 4, 2014
|
|
5KSW
 
 | | DHODB-I74D mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2016-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Isoleucine 74 Plays a Key Role in Controlling Electron Transfer Between Lactococcus lactis Dihydroorotate Dehydrogenase 1B Subunits
To Be Published
|
|
4M5P
 
 | | OYE2.6 Y78W, I113C | | Descriptor: | FLAVIN MONONUCLEOTIDE, MALONIC ACID, NADPH dehydrogenase, ... | | Authors: | Sullivan, B, Stewart, J.D. | | Deposit date: | 2013-08-08 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Residues Controlling Facial Selectivity in an Alkene Reductase and Semirational Alterations to Create Stereocomplementary Variants.
ACS catalysis, 4, 2014
|
|
4YNC
 
 | | OYE1 W116A COMPLEXED WITH (Z)-METHYL-3-CYANO-3-PHENYLACRYLATE IN A NON PRODUCTIVE BINDING MODE | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, ... | | Authors: | Santangelo, S, Brenna, E, Stewart, J.D, Powell III, R.W. | | Deposit date: | 2015-03-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Opposite Enantioselectivity in the Bioreduction of (Z)-beta-Aryl-beta-cyanoacrylates Mediated by the Tryptophan 116 Mutants of Old Yellow Enzyme 1: Synthetic Approach to (R)- and (S)-beta-Aryl-gamma-lactams
Adv.Synth.Catal., 357, 2015
|
|
4YIL
 
 | | OYE1 W116A COMPLEXED WITH (Z)-METHYL 3-CYANO-3-(4-FLUOROPHENYL)ACRYLATE IN A NON PRODUCTIVE BINDING MODE | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, NADPH dehydrogenase 1, ... | | Authors: | Santangelo, S, Brenna, E, Stewart, J.D, Powell III, R.W. | | Deposit date: | 2015-03-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Opposite Enantioselectivity in the Bioreduction of (Z)-beta-Aryl-beta-cyanoacrylates Mediated by the Tryptophan 116 Mutants of Old Yellow Enzyme 1: Synthetic Approach to (R)- and (S)-beta-Aryl-gamma-lactams
Adv.Synth.Catal., 357, 2015
|
|
1JQL
 
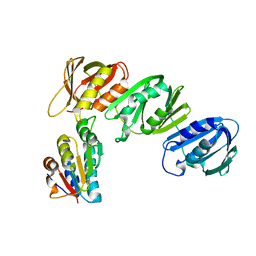 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of beta-delta (1-140) | | Descriptor: | DNA Polymerase III, BETA CHAIN, DELTA SUBUNIT | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JQJ
 
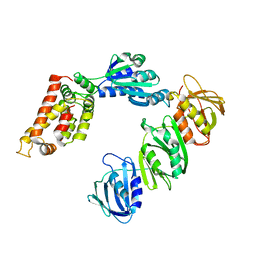 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of the beta-delta complex | | Descriptor: | DNA polymerase III, beta chain, delta subunit | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
5VYE
 
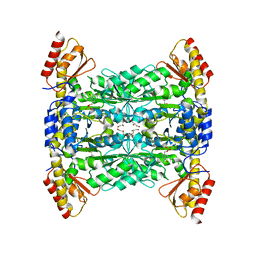 | |
4K7V
 
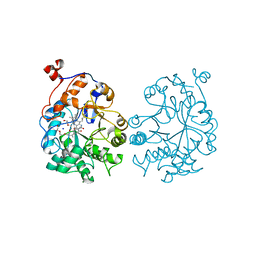 | | OYE1-W116A complexed with (R)-carvone | | Descriptor: | (5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sullivan, B, Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the
Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
5UE9
 
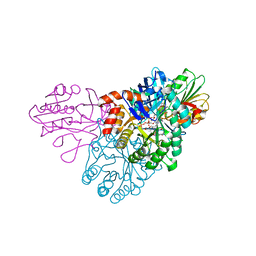 | | WT DHODB with orotate bound | | Descriptor: | CHLORIDE ION, Dihydroorotate dehydrogenase, Dihydroorotate dehydrogenase B (NAD(+)), ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Isoleucine 74 Plays a Key Role in Controlling Electron Transfer Between Lactococcus lactis Dihydroorotate Dehydrogenase 1B Subunits
to be published
|
|
4GBU
 
 | |
4GE8
 
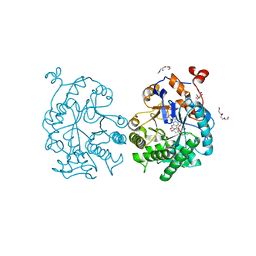 | | OYE1-W116I complexed with (s)-Carvone | | Descriptor: | (5S)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
4GWE
 
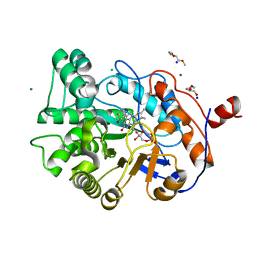 | | W116L-OYE1 complexed with (R)-carvone | | Descriptor: | (5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2012-09-02 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
4GXM
 
 | |
4H4I
 
 | |
4K7Y
 
 | | Oye1-w116t | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sullivan, B, Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the
Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
4K8H
 
 | | OYE1-W116V complexed with (R)-carvone | | Descriptor: | (5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sullivan, B, Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the
Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
4K8E
 
 | | OYE1-W116V complexed with the aromatic product of (R)-carvone dismutation | | Descriptor: | 2-methyl-5-(prop-1-en-2-yl)phenol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sullivan, B, Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the
Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
4H6K
 
 | | W116I mutant of OYE1 | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the
Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
3TJL
 
 | |
3UPW
 
 | | Pichia Stipitis OYE2.6 complexed with nicotinamide | | Descriptor: | FLAVIN MONONUCLEOTIDE, MALONIC ACID, NICOTINAMIDE, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2011-11-18 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Structural and Catalytic Characterization of Pichia stipitis OYE 2.6, a Useful Biocatalyst for Asymmetric Alkene Reductions
Adv.Synth.Catal., 2012
|
|
4DF2
 
 | | P. stipitis OYE2.6 complexed with p-Chlorophenol | | Descriptor: | 4-chlorophenol, FLAVIN MONONUCLEOTIDE, MALONIC ACID, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2012-01-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Structural and Catalytic Characterization of Pichia stipitis OYE 2.6, a Useful Biocatalyst for Asymmetric Alkene Reductions
Adv.Synth.Catal., 2012
|
|
