1J6S
 
 | | Crystal Structure of an RNA Tetraplex (UGAGGU)4 with A-tetrads, G-tetrads, U-tetrads and G-U octads | | Descriptor: | 5'-R(*(BRUP*GP*AP*GP*GP*U)-3', BARIUM ION, SODIUM ION | | Authors: | Pan, B, Xiong, Y, Shi, K, Deng, J, Sundaralingam, M. | | Deposit date: | 2002-07-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads:
implications for specific binding in RNA tetraplexes
Structure, 11, 2003
|
|
6UPS
 
 | |
4ZWG
 
 | | Crystal structure of the GTP-dATP-bound catalytic core of SAMHD1 phosphomimetic T592E mutant | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tang, C, Ji, X, Xiong, Y. | | Deposit date: | 2015-05-19 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impaired dNTPase Activity of SAMHD1 by Phosphomimetic Mutation of Thr-592.
J.Biol.Chem., 290, 2015
|
|
6X1H
 
 | |
4ZWE
 
 | |
3OVA
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CCA-adding enzyme, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
6X1G
 
 | |
6U6V
 
 | | Crystal structure of human PD-1H / VISTA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | Authors: | Slater, B.T, Han, X, Chen, L, Xiong, Y. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into T cell coinhibition by PD-1H (VISTA).
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7N9X
 
 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | Descriptor: | Capsid protein, Nanobody, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.511 Å) | | Cite: | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
6OBH
 
 | | Structure of HIV-1 CA 1/2-hexamer | | Descriptor: | CA, SODIUM ION | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
3OVS
 
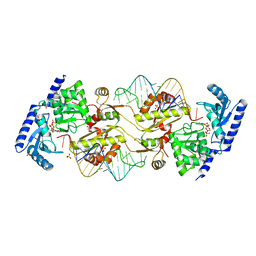 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3MYZ
 
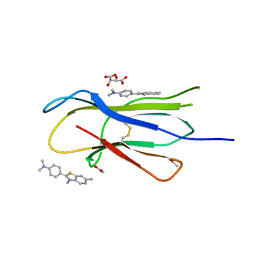 | | Protein induced photophysical changes to the amyloid indicator dye, thioflavin T | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Wolfe, L.S, Calabrese, M.F, Nath, A, Blaho, D.V, Miranker, A.D, Xiong, Y. | | Deposit date: | 2010-05-11 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein-induced photophysical changes to the amyloid indicator dye thioflavin T.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MZT
 
 | | Protein-induced photophysical changes to the amyloid indicator dye, thioflavin T | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Beta-2-microglobulin, COPPER (II) ION | | Authors: | Wolfe, L.S, Calabrese, M.F, Nath, A, Blaho, D.V, Miranker, A.D, Xiong, Y. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein-induced photophysical changes to the amyloid indicator dye thioflavin T.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OVB
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3OUY
 
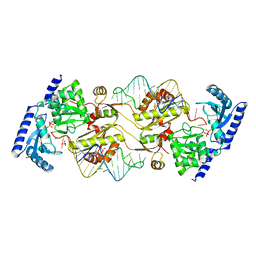 | | How the CCA-adding Enzyme Selects Adenine Over Cytosine at Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CCA-Adding Enzyme, PYROPHOSPHATE 2-, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3OV7
 
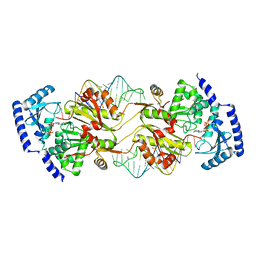 | | How the CCA-Adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3P49
 
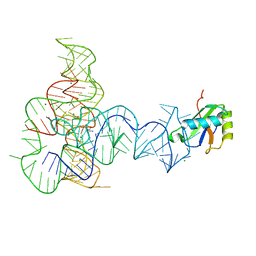 | | Crystal Structure of a Glycine Riboswitch from Fusobacterium nucleatum | | Descriptor: | GLYCINE, GLYCINE RIBOSWITCH, MAGNESIUM ION, ... | | Authors: | Butler, E.B, Wang, J, Xiong, Y, Strobel, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of cooperative ligand binding by the glycine riboswitch.
Chem.Biol., 18, 2011
|
|
1MDG
 
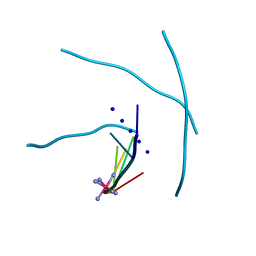 | | An Alternating Antiparallel Octaplex in an RNA Crystal Structure | | Descriptor: | 5'-R(*UP*(BGM)GP*AP*GP*GP*U)-3', COBALT HEXAMMINE(III), SODIUM ION | | Authors: | Pan, B.C, Xiong, Y, Shi, K, Sundaralingam, M. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Eight-Stranded Helical Fragment in RNA Crystal Structure: Implications for Tetraplex Interaction
Structure, 11, 2003
|
|
5TTF
 
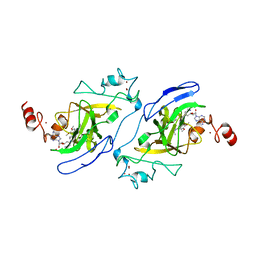 | | Crystal structure of catalytic domain of G9a with MS012 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EHMT2, N4-(1-methylpiperidin-4-yl)-N2-hexyl-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5TTG
 
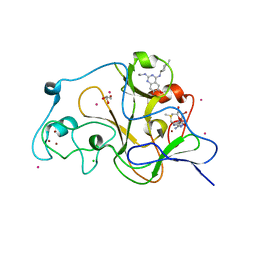 | | Crystal structure of catalytic domain of GLP with MS012 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5V9J
 
 | | Crystal structure of catalytic domain of GLP with MS0105 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of GLP with MS0105
to be published
|
|
1YQY
 
 | | Structure of B. Anthrax Lethal factor in complex with a hydroxamate inhibitor | | Descriptor: | (2R)-2-{[(4-FLUORO-3-METHYLPHENYL)SULFONYL]AMINO}-N-HYDROXY-2-TETRAHYDRO-2H-PYRAN-4-YLACETAMIDE, Lethal factor, ZINC ION | | Authors: | Shoop, W.L, Xiong, Y, Wiltsie, J, Woods, A, Guo, J, Pivnichny, J.V, Felcetto, T, Michael, B.F, Bansal, A. | | Deposit date: | 2005-02-02 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anthrax lethal factor inhibition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5V9I
 
 | | Crystal structure of catalytic domain of G9a with MS0105 | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EHMT2, N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of G9a with MS0105
to be published
|
|
1U6G
 
 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
4REC
 
 | | A nuclease-DNA complex form 3 | | Descriptor: | DNA (40-MER), Fanconi-associated nuclease 1, IODIDE ION | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
