4W5U
 
 | |
4V0I
 
 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4V1N
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4UXV
 
 | | Cytoplasmic domain of bacterial cell division protein EzrA | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
5P21
 
 | |
8BTM
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GdmF | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
8JP3
 
 | | FCP trimer in diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | Authors: | Feng, Y, Li, Z, Zhou, C.C, Liu, C, Shen, J.R, Wang, W. | | Deposit date: | 2023-06-10 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | FCP trimer in diatom Thalassiosira pseudonana
To Be Published
|
|
8T15
 
 | |
8T17
 
 | |
8T18
 
 | |
4UXL
 
 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2014-08-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5OQK
 
 | | Solution NMR structure of truncated, human Hv1/VSOP (Voltage-gated proton channel) | | Descriptor: | Voltage-gated hydrogen channel 1 | | Authors: | Bayrhuber, M, Maslennikov, I, Kwiatowski, W, Sobol, A, Wierschem, C, Eichmann, C, Riek, R. | | Deposit date: | 2017-08-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure and Functional Behavior of the Human Proton Channel.
Biochemistry, 58, 2019
|
|
8C3D
 
 | | Sulfonated Calpeptin is a promising drug candidate against SARS-CoV-2 infections | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CALCIUM ION, Cathepsin K | | Authors: | Loboda, J, Karnicar, K, Lindic, N, Usenik, A, Lieske, J, Meents, A, Guenther, S, Reinke, P.Y.A, Falke, S, Ewert, W, Turk, D. | | Deposit date: | 2022-12-23 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
1FIA
 
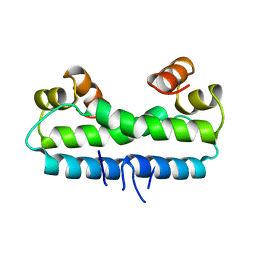 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
1FBZ
 
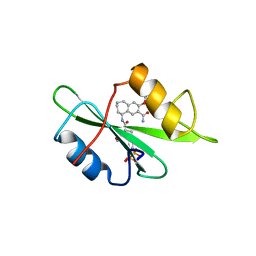 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | Authors: | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
7NXF
 
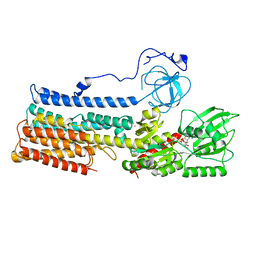 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
7NY1
 
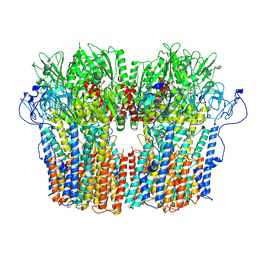 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
1FGT
 
 | |
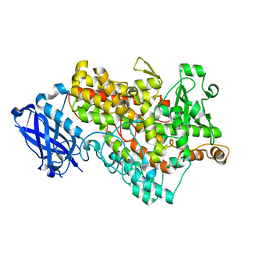
1F8N
 
 | |
1FPH
 
 | |
1FGQ
 
 | |
1FI1
 
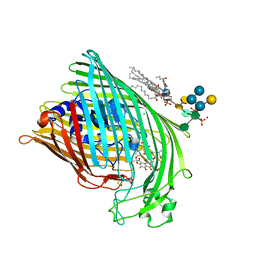 | | FhuA in complex with lipopolysaccharide and rifamycin CGP4832 | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Koedding, J, Boes, C, Walker, G, Coulton, J.W, Diederichs, K, Braun, V, Welte, W. | | Deposit date: | 2000-08-03 | | Release date: | 2001-08-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA.
Structure, 9, 2001
|
|
1FGR
 
 | |
5OQV
 
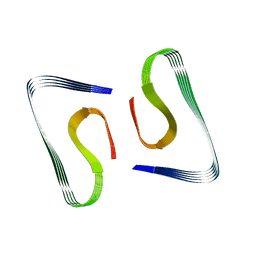 | | Near-atomic resolution fibril structure of complete amyloid-beta(1-42) by cryo-EM | | Descriptor: | Amyloid beta A4 protein | | Authors: | Gremer, L, Schoelzel, D, Schenk, C, Reinartz, E, Labahn, J, Ravelli, R, Tusche, M, Lopez-Iglesias, C, Hoyer, W, Heise, H, Willbold, D, Schroeder, G.F. | | Deposit date: | 2017-08-14 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Fibril structure of amyloid-beta (1-42) by cryo-electron microscopy.
Science, 358, 2017
|
|
5OWD
 
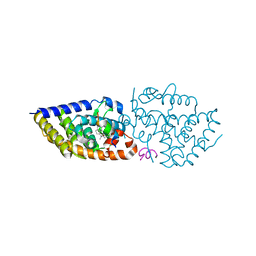 | | Vitamin D receptor complex | | Descriptor: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-7~{a}-methyl-1-[(2~{S})-6-methyl-2-oxidanyl-hept-5-en-2-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
