1LI1
 
 | | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link | | Descriptor: | ACETATE ION, Collagen alpha 1(IV), Collagen alpha 2(IV) | | Authors: | Than, M.E, Henrich, S, Huber, R, Ries, A, Mann, K, Kuhn, K, Timpl, R, Bourenkov, G.P, Bartunik, H.D, Bode, W. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1C52
 
 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | Descriptor: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | Deposit date: | 1997-06-23 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
1S4V
 
 | | The 2.0 A crystal structure of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm | | Descriptor: | DVA-LEU-LYS-0QE peptide, SULFATE ION, cysteine endopeptidase | | Authors: | Than, M.E, Helm, M, Simpson, D.J, Lottspeich, F, Huber, R, Gietl, C. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm.
J.Mol.Biol., 336, 2004
|
|
5JXI
 
 | | Structure of the unliganded form of the proprotein convertase furin in presence of EDTA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXG
 
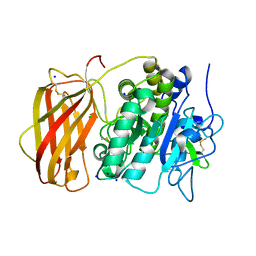 | | Structure of the unliganded form of the proprotein convertase furin. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3KTM
 
 | | Structure of the Heparin-induced E1-Dimer of the Amyloid Precursor Protein (APP) | | Descriptor: | (3R)-butane-1,3-diol, ACETATE ION, Amyloid beta A4 protein, ... | | Authors: | Dahms, S.O, Hoefgen, S, Roeser, D, Schlott, B, Guhrs, K.H, Than, M.E. | | Deposit date: | 2009-11-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2F9N
 
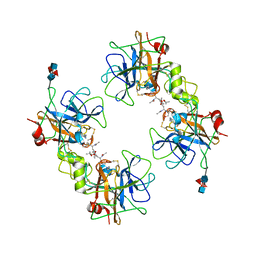 | | Crystal Structure of the Recombinant Human Alpha I Tryptase Mutant K192Q/D216G in Complex with Leupeptin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Leupeptin, ... | | Authors: | Rohr, K.B, Selwood, T, Marquardt, U, Huber, R, Schechter, N.M, Bode, W, Than, M.E. | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray Structures of Free and Leupeptin-complexed Human alpha I-Tryptase Mutants: Indication for an alpha to beta-Tryptase Transition
J.Mol.Biol., 357, 2005
|
|
2F9P
 
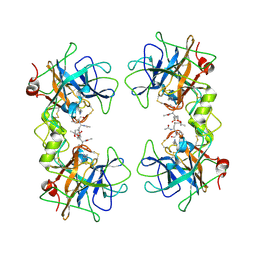 | | Crystal Structure of the Recombinant Human Alpha I Tryptase Mutant D216G in Complex with Leupeptin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Leupeptin, ... | | Authors: | Rohr, K.B, Selwood, T, Marquardt, U, Huber, R, Schechter, N.M, Bode, W, Than, M.E. | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structures of Free and Leupeptin-complexed Human alpha I-Tryptase Mutants: Indication for an alpha to beta-Tryptase Transition
J.Mol.Biol., 357, 2005
|
|
2F9O
 
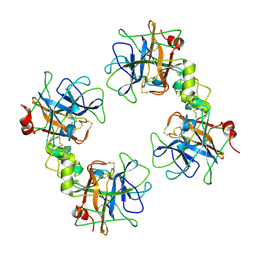 | | Crystal Structure of the Recombinant Human Alpha I Tryptase Mutant D216G | | Descriptor: | Tryptase alpha-1, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Rohr, K.B, Selwood, T, Marquardt, U, Huber, R, Schechter, N.M, Bode, W, Than, M.E. | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of Free and Leupeptin-complexed Human alpha I-Tryptase Mutants: Indication for an alpha to beta-Tryptase Transition
J.Mol.Biol., 357, 2005
|
|
6EQX
 
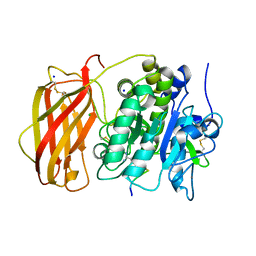 | |
6EQW
 
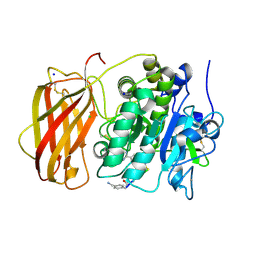 | |
5JMO
 
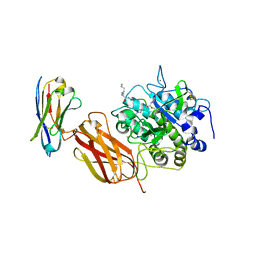 | |
6EQV
 
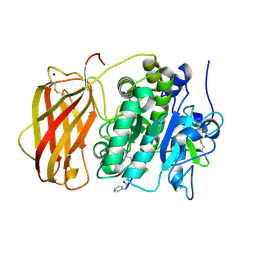 | |
5JXH
 
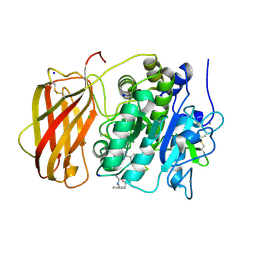 | | Structure the proprotein convertase furin in complex with meta-guanidinomethyl-Phac-RVR-Amba at 2.0 Angstrom resolution. | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JMR
 
 | | X-ray structure of the furin inhibitory antibody Nb14 | | Descriptor: | CHLORIDE ION, camelid VHH fragment | | Authors: | Dahms, S.O, Than, M.E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers.
Sci Rep, 6, 2016
|
|
5JXJ
 
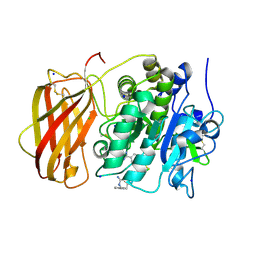 | | Structure of the proprotein convertase furin complexed to meta-guanidinomethyl-Phac-RVR-Amba in presence of EDTA | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1B33
 
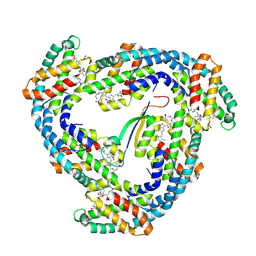 | | STRUCTURE OF LIGHT HARVESTING COMPLEX OF ALLOPHYCOCYANIN ALPHA AND BETA CHAINS/CORE-LINKER COMPLEX AP*LC7.8 | | Descriptor: | ALLOPHYCOCYANIN, ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Reuter, W, Wiegand, G, Huber, R, Than, M.E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis at 2.2 A of orthorhombic crystals presents the asymmetry of the allophycocyanin-linker complex, AP.LC7.8, from phycobilisomes of Mastigocladus laminosus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5MIM
 
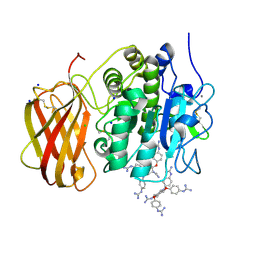 | | Xray structure of human furin bound with the 2,5-dideoxystreptamine derived small molecule inhibitor 1n | | Descriptor: | 1-[(1~{R},2~{R},4~{S},5~{S})-2,4-bis(4-carbamimidamidophenoxy)-5-[(4-carbamimidamidophenyl)amino]cyclohexyl]guanidine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Guan-Sheng, J, Than, M.E. | | Deposit date: | 2016-11-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies Revealed Active Site Distortions of Human Furin by a Small Molecule Inhibitor.
ACS Chem. Biol., 12, 2017
|
|
1C6O
 
 | | CRYSTAL STRUCTURE OF OXIDIZED CYTOCHROME C6 FROM THE GREEN ALGAE SCENEDESMUS OBLIQUUS | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schnackenberg, J, Than, M.E, Mann, K, Wiegand, G, Huber, R, Reuter, W. | | Deposit date: | 1999-04-07 | | Release date: | 2000-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino acid sequence, crystallization and structure determination of reduced and oxidized cytochrome c6 from the green alga Scenedesmus obliquus.
J.Mol.Biol., 290, 1999
|
|
1C6R
 
 | | CRYSTAL STRUCTURE OF REDUCED CYTOCHROME C6 FROM THE GREEN ALGAE SCENEDESMUS OBLIQUUS | | Descriptor: | CADMIUM ION, CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schnackenberg, J, Than, M.E, Mann, K, Wiegand, G, Huber, R, Reuter, W. | | Deposit date: | 1999-04-06 | | Release date: | 2000-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amino acid sequence, crystallization and structure determination of reduced and oxidized cytochrome c6 from the green alga Scenedesmus obliquus.
J.Mol.Biol., 290, 1999
|
|
3QO4
 
 | | The Crystal Structure of Death Receptor 6 | | Descriptor: | ACETATE ION, SULFATE ION, Tumor necrosis factor receptor superfamily member 21 | | Authors: | Kuester, M, Kemmerzehl, S, Dahms, S.O, Roeser, D, Than, M.E. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-18 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of death receptor 6 (DR6): a potential receptor of the amyloid precursor protein (APP).
J.Mol.Biol., 409, 2011
|
|
3UMH
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with cadmium | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
3UMI
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with zinc | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
3UMK
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with copper | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
1P8J
 
 | | CRYSTAL STRUCTURE OF THE PROPROTEIN CONVERTASE FURIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DECANOYL-ARG-VAL-LYS-ARG-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Henrich, S, Cameron, A, Bourenkov, G.P, Kiefersauer, R, Huber, R, Lindberg, I, Bode, W, Than, M.E. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Proprotein Processing Proteinase Furin Explains its Stringent Specificity
Nat.Struct.Biol., 10, 2003
|
|
