6GN5
 
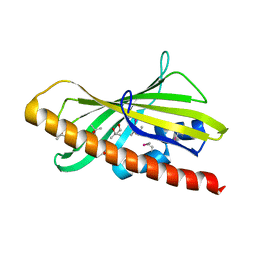 | | CRYSTAL STRUCTURE OF HUMAN GRAMD1C START DOMAIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GRAM domain-containing protein 1C | | Authors: | Friese, A, Vetter, I.R. | | Deposit date: | 2018-05-30 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis.
Nat.Chem.Biol., 15, 2019
|
|
4BQD
 
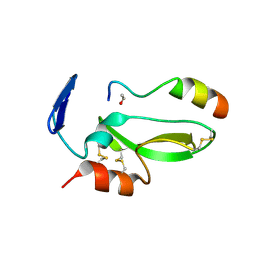 | | KD1 of human TFPI in complex with a synthetic peptide | | Descriptor: | GLYCEROL, PEPTIDE, TISSUE FACTOR PATHWAY INHIBITOR (LIPOPROTEIN-ASSOCIATED COAGULATION INHIBITOR) VARIANT | | Authors: | Griessner, A, Brandstetter, H. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Small Peptides Blocking Inhibition of Factor Xa and Tissue Factor-Factor Viia by Tissue Factor Pathway Inhibitor (Tfpi)
J.Biol.Chem., 289, 2014
|
|
1LI1
 
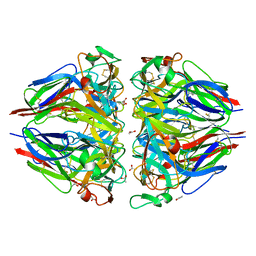 | | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link | | Descriptor: | ACETATE ION, Collagen alpha 1(IV), Collagen alpha 2(IV) | | Authors: | Than, M.E, Henrich, S, Huber, R, Ries, A, Mann, K, Kuhn, K, Timpl, R, Bourenkov, G.P, Bartunik, H.D, Bode, W. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7O5H
 
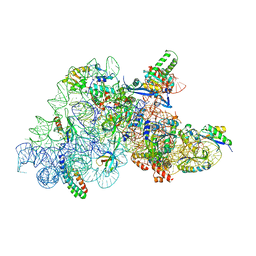 | | Ribosomal methyltransferase KsgA bound to small ribosomal subunit | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Stephan, N.C, Ries, A.B, Boehringer, D, Ban, N. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-16 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of successive adenosine modifications by the conserved ribosomal methyltransferase KsgA.
Nucleic Acids Res., 49, 2021
|
|
1H4U
 
 | | Domain G2 of mouse nidogen-1 | | Descriptor: | NIDOGEN-1 | | Authors: | Hopf, M, Gohring, W, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure and Mutational Analysis of a Perlecan-Binding Fragment of Nidogen-1
Nat.Struct.Biol., 8, 2001
|
|
1GL4
 
 | | Nidogen-1 G2/Perlecan IG3 Complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BASEMENT MEMBRANE-SPECIFIC HEPARAN SULFATE PROTEOGLYCAN CORE PROTEIN, NIDOGEN-1, ... | | Authors: | Kvansakul, M, Hopf, M, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the High-Affinity Interaction of Nidogen-1 with Immunoglobulin-Like Domain 3 of Perlecan
Embo J., 20, 2001
|
|
1C1Z
 
 | | CRYSTAL STRUCTURE OF HUMAN BETA-2-GLYCOPROTEIN-I (APOLIPOPROTEIN-H) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA2-GLYCOPROTEIN-I, ... | | Authors: | Schwarzenbacher, R, Zeth, K, Diederichs, K, Gries, A, Kostner, G.M, Laggner, P, Prassl, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome.
EMBO J., 18, 1999
|
|
6Y9S
 
 | | Crystal structure of GSK-3b in complex with the imidazo[1,5-a]pyridine-3-carboxamide inhibitor 16 | | Descriptor: | ACETATE ION, Glycogen synthase kinase-3 beta, ~{N}-(oxan-4-ylmethyl)-6-(5-propan-2-yloxypyridin-3-yl)imidazo[1,5-a]pyridine-3-carboxamide | | Authors: | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
6Y9R
 
 | | Crystal structure of GSK-3b in complex with the 1H-indazole-3-carboxamide inhibitor 2 | | Descriptor: | ACETATE ION, GLYCEROL, Glycogen synthase kinase-3 beta, ... | | Authors: | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
6TPF
 
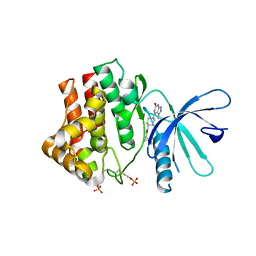 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | (1~{S})-2,2-bis(fluoranyl)-~{N}-[4-(3-methyl-6-oxidanylidene-2,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]cyclopropane-1-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Griessner, A, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
6HGY
 
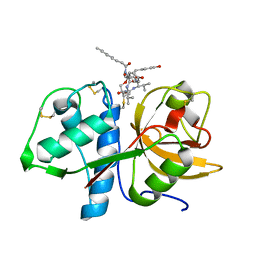 | | CRYSTAL STRUCTURE OF CATHEPSIN K WITH N-DESMETHYL THALASSOSPIRAMIDE C | | Descriptor: | Cathepsin K, THALASSOSPIRAMIDE C | | Authors: | Zakarian, A, Buckman, B.O, Adler, M, Griessner, A, Blaesse, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Total Synthesis of Covalent Cysteine Protease Inhibitor N-Desmethyl Thalassospiramide C and Crystallographic Evidence for Its Mode of Action.
Org.Lett., 21, 2019
|
|
6S41
 
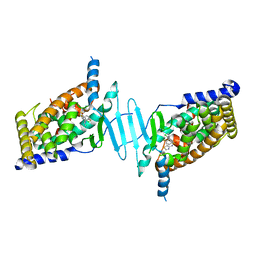 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH XPC-7455 | | Descriptor: | 4-[[(1~{S})-1-[2,5-bis(fluoranyl)phenyl]ethyl]amino]-5-chloranyl-2-fluoranyl-~{N}-(1,3-thiazol-4-yl)benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Focken, T, Maskos, K, Griessner, A, Krapp, S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of CNS-Penetrant Aryl Sulfonamides as Isoform-Selective NaV1.6 Inhibitors with Efficacy in Mouse Models of Epilepsy.
J.Med.Chem., 62, 2019
|
|
3F8C
 
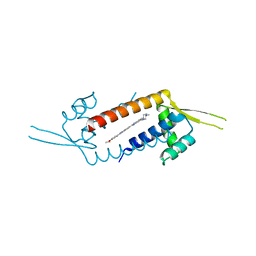 | | Crystal structure of multidrug binding transcriptional regulator LmrR complexed with Hoechst 33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
3F8B
 
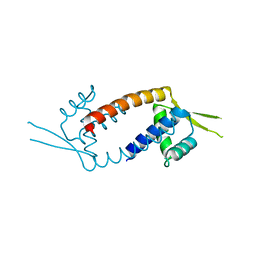 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in drug free state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
3F8F
 
 | | Crystal structure of multidrug binding transcriptional regulator LmrR complexed with Daunomycin | | Descriptor: | DAUNOMYCIN, Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
1OXV
 
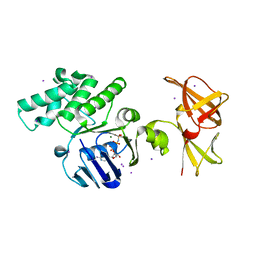 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION, ... | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus:
nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
1OXU
 
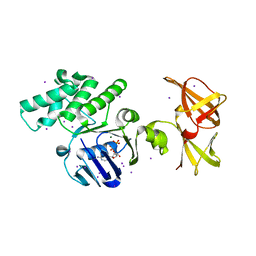 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
1OXT
 
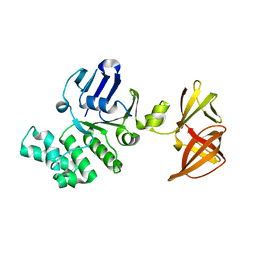 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus:
nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
1OXS
 
 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus:
nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
1UCS
 
 | | Type III Antifreeze Protein RD1 from an Antarctic Eel Pout | | Descriptor: | Antifreeze peptide RD1 | | Authors: | Ko, T.-P, Robinson, H, Gao, Y.-G, Cheng, C.-H.C, DeVries, A.L, Wang, A.H.-J. | | Deposit date: | 2003-04-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.62 Å) | | Cite: | The refined crystal structure of an eel pout type III antifreeze protein RD1 at 0.62-A resolution reveals structural microheterogeneity of protein and solvation.
Biophys.J., 84, 2003
|
|
1G4I
 
 | | Crystal structure of the bovine pancreatic phospholipase A2 at 0.97A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Steiner, R.A, Rozeboom, H.J, de Vries, A, Kalk, K.H, Murshudov, G.N, Wilson, K.S, Dijkstra, B.W. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | X-ray structure of bovine pancreatic phospholipase A2 at atomic resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1OXX
 
 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.-V, van Oosterwijk, N, Dijkstra, B.W, Driessen, A.J.M, Thunnissen, A.M.W.H. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus
J.Mol.Biol., 334, 2003
|
|
