1OK1
 
 | | Decay accelerating factor (cd55) : the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8GEK
 
 | | Dihydrodipicolinate synthase with pyruvate from Candidatus Liberibacter solanacearum | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase | | Authors: | Gilkes, J.M, Frampton, R.A, Board, A, Sheen, C.R, Smith, G.R, Dobson, R.C.J. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dihydrodipicolinate synthase with pyruvate from the plant pathogen, Candidatus Liberibacter solanacearum
To Be Published
|
|
6UT2
 
 | | 3D structure of the leiomodin/tropomyosin binding interface | | Descriptor: | Leiomodin-2, Tropomyosin alpha-1 chain chimeric peptide | | Authors: | Tolkatchev, D, Smith, G.E, Helms, G.L, Cort, J.R, Kostyukova, A.S. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Leiomodin creates a leaky cap at the pointed end of actin-thin filaments.
Plos Biol., 18, 2020
|
|
1LPH
 
 | | LYS(B28)PRO(B29)-HUMAN INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Ciszak, E, Beals, J.M, Frank, B.H, Baker, J.C, Carter, N.D, Smith, G.D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-06-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin.
Structure, 3, 1995
|
|
1TRZ
 
 | |
3EOC
 
 | | Cdk2/CyclinA complexed with a imidazo triazin-2-amine | | Descriptor: | 5-methyl-7-phenyl-N-(3,4,5-trimethoxyphenyl)imidazo[5,1-f][1,2,4]triazin-2-amine, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Cheung, M, Kuntz, K, Pobanz, M, Salovich, J, Wilson, B, Andrews, W, Shewchuk, L, Epperly, A, Hassler, D, Leesnitzer, M, Smith, J, Smith, G, Lansing, T, Mook, R. | | Deposit date: | 2008-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Imidazo[5,1-f][1,2,4]triazin-2-amines as novel inhibitors of polo-like kinase 1.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1B1C
 
 | | CRYSTAL STRUCTURE OF THE FMN-BINDING DOMAIN OF HUMAN CYTOCHROME P450 REDUCTASE AT 1.93A RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH-CYTOCHROME P450 REDUCTASE) | | Authors: | Zhao, Q, Modi, S, Smith, G, Paine, M, Mcdonagh, P.D, Wolf, C.R, Tew, D, Lian, L.-Y, Roberts, G.C.K, Driessen, H.P.C. | | Deposit date: | 1998-11-19 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution.
Protein Sci., 8, 1999
|
|
7LOY
 
 | | Dihydrodipicolinate synthase with pyruvate from Candidatus Liberibacter solanacearum | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase | | Authors: | Gilkes, J.M, Frampton, R.A, Board, A, Sheen, C.R, Smith, G.R, Dobson, R.C.J. | | Deposit date: | 2021-02-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dihydrodipicolinate synthase with pyruvate from the plant pathogen, Candidatus Liberibacter solanacearum
To Be Published
|
|
7LVL
 
 | | Dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine from Candidatus Liberibacter solanacearum | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, LYSINE | | Authors: | Gilkes, J.M, Frampton, R.A, Board, A.J, Sheen, C.R, Smith, G.R, Dobson, R.C.J.D. | | Deposit date: | 2021-02-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine from Candidatus Liberibacter solanacearum
To Be Published
|
|
7MJF
 
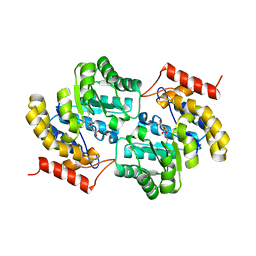 | | Crystal structure of Candidatus Liberibacter solanacearum dihydrodipicolinate synthase with pyruvate and succinic semi-aldehyde bound in active site | | Descriptor: | (4R)-4-oxidanyl-2-oxidanylidene-heptanedioic acid, (4S)-4-hydroxy-2-oxoheptanedioic acid, 4-hydroxy-tetrahydrodipicolinate synthase | | Authors: | Gilkes, J, Frampton, R.A, Board, A.J, Sheen, C.R, Smith, G.R, Dobson, R.C.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Candidatus Liberibacter solanacearum dihydrodipicolinate synthase with pyruvate and succinic semi-aldehyde bound in active site
To Be Published
|
|
1FU2
 
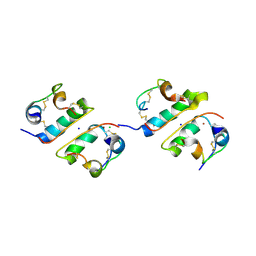 | | FIRST PROTEIN STRUCTURE DETERMINED FROM X-RAY POWDER DIFFRACTION DATA | | Descriptor: | CHLORIDE ION, INSULIN, A CHAIN, ... | | Authors: | Von Dreele, R.B, Stephens, P.W, Blessing, R.H, Smith, G.D. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-16 | | Last modified: | 2018-10-03 | | Method: | POWDER DIFFRACTION | | Cite: | The first protein crystal structure determined from high-resolution X-ray powder diffraction data: a variant of T3R3 human insulin-zinc complex produced by grinding.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1Z5N
 
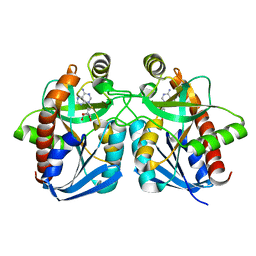 | | Crystal structure of MTA/AdoHcy nucleosidase Glu12Gln mutant complexed with 5-methylthioribose and adenine | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
4D5T
 
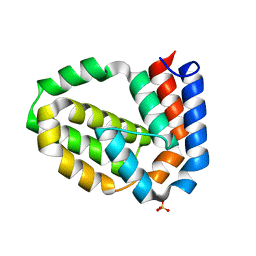 | | Structure of N-terminally truncated A49 from Vaccinia Virus Western Reserve | | Descriptor: | PROTEIN A49R, SULFATE ION | | Authors: | Neidel, S, Maluquer de Motes, C, Mansur, D.S, Strnadova, P, Smith, G.L, Graham, S.C. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Vaccinia Virus Protein A49 is an Unexpected Member of the B-Cell Lymphoma (Bcl)-2 Protein Family
J.Biol.Chem., 290, 2015
|
|
1TJV
 
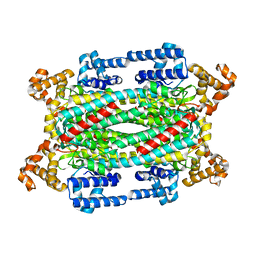 | | Crystal Structure of T161D Duck Delta 2 Crystallin Mutant | | Descriptor: | Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
BIOCHEM.J., 384, 2004
|
|
1TJU
 
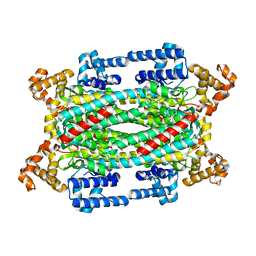 | | Crystal Structure of T161S Duck Delta 2 Crystallin Mutant | | Descriptor: | Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
Biochem.J., 384, 2004
|
|
1TJW
 
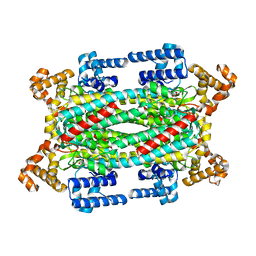 | | Crystal Structure of T161D Duck Delta 2 Crystallin Mutant with bound argininosuccinate | | Descriptor: | ARGININOSUCCINATE, Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
Biochem.J., 384, 2004
|
|
1FUB
 
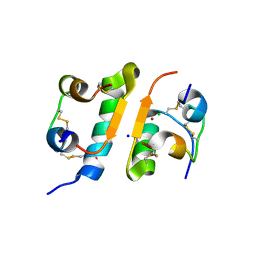 | | FIRST PROTEIN STRUCTURE DETERMINED FROM X-RAY POWDER DIFFRACTION DATA | | Descriptor: | CHLORIDE ION, INSULIN, A CHAIN, ... | | Authors: | Von Dreele, R.B, Stephens, P.W, Blessing, R.H, Smith, G.D. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-16 | | Last modified: | 2018-10-03 | | Method: | POWDER DIFFRACTION | | Cite: | The first protein crystal structure determined from high-resolution X-ray powder diffraction data: a variant of T3R3 human insulin-zinc complex produced by grinding.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4BBC
 
 | | THE STRUCTURE OF VACCINIA VIRUS N1 R71Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
4BBB
 
 | | THE STRUCTURE OF VACCINIA VIRUS N1 Q61Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
4BBD
 
 | | THE STRUCTURE OF VACCINIA VIRUS N1 R58Y MUTANT | | Descriptor: | N1L | | Authors: | Maluquer de Motes, C, Cooray, S, McGourty, K, Ren, H, Bahar, M.W, Stuart, D.I, Grimes, J.M, Graham, S.C, Smith, G.L. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of Apoptosis and NF-kappaB Activation by Vaccinia Protein N1 Occur Via Distinct Binding Surfaces and Make Different Contributions to Virulence.
Plos Pathog., 7, 2011
|
|
1NOQ
 
 | | e-motif structure | | Descriptor: | 5'-D(*CP*CP*GP*CP*CP*G)-3' | | Authors: | Zheng, M, Huang, X, Smith, G.K, Yang, X, Gao, X. | | Deposit date: | 2003-01-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding.
An NMR and UV spectroscopic study.
J.Mol.Biol., 264, 1996
|
|
1Z5P
 
 | | Crystal structure of MTA/AdoHcy nucleosidase with a ligand-free purine binding site | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5O
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Asp197Asn mutant complexed with 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
3BL6
 
 | | Crystal structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Siu, K.K.W, Lee, J.E, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1A7A
 
 | | STRUCTURE OF HUMAN PLACENTAL S-ADENOSYLHOMOCYSTEINE HYDROLASE: DETERMINATION OF A 30 SELENIUM ATOM SUBSTRUCTURE FROM DATA AT A SINGLE WAVELENGTH | | Descriptor: | (1'R,2'S)-9-(2-HYDROXY-3'-KETO-CYCLOPENTEN-1-YL)ADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Turner, M.A, Yuan, C.-S, Borchardt, R.T, Hershfield, M.S, Smith, G.D, Howell, P.L. | | Deposit date: | 1998-03-10 | | Release date: | 1999-04-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength.
Nat.Struct.Biol., 5, 1998
|
|
