3C0J
 
 | |
3DAQ
 
 | | Crystal structure of dihydrodipicolinate synthase from methicillin-resistant Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Dobson, R.C.J, Burgess, B.R, Jameson, G.B, Gerrard, J.A, Parker, M.W, Perugini, M.A. | | Deposit date: | 2008-05-29 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and evolution of a novel dimeric enzyme from a clinically-important bacterial pathogen.
J.Biol.Chem., 2008
|
|
1S5T
 
 | |
1S5V
 
 | |
1S5W
 
 | |
3DU0
 
 | | E. coli dihydrodipicolinate synthase with first substrate, pyruvate, bound in active site | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Dobson, R.C.J, Devenish, S.R.A, Gerrard, J.A, Jameson, G.B. | | Deposit date: | 2008-07-16 | | Release date: | 2008-11-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The high-resolution structure of dihydrodipicolinate synthase from Escherichia coli bound to its first substrate, pyruvate.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3IRD
 
 | |
3QGU
 
 | | L,L-Diaminopimelate aminotransferase from Chlamydomonas reinhardtii | | Descriptor: | AZIDE ION, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Dobson, R.C.J, Giron, I, Hudson, A.O. | | Deposit date: | 2011-01-25 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | L,L-Diaminopimelate Aminotransferase from Chlamydomonas reinhardtii: A Target for Algaecide Development
Plos One, 6, 2011
|
|
1YXC
 
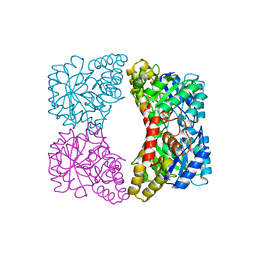 | | Structure of E. coli dihydrodipicolinate synthase to 1.9 A | | Descriptor: | CHLORIDE ION, POTASSIUM ION, dihydrodipicolinate synthase | | Authors: | Dobson, R.C.J, Griffin, M.D.W, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-02-20 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of native and (S)-lysine-bound dihydrodipicolinate synthase from Escherichia coli with improved resolution show new features of biological significance.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YXD
 
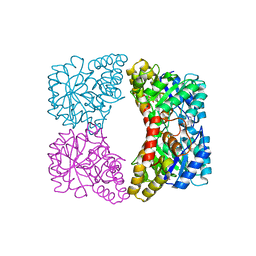 | | Structure of E. coli dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine to 2.0 A | | Descriptor: | CHLORIDE ION, LYSINE, POTASSIUM ION, ... | | Authors: | Dobson, R.C.J, Griffin, M.D.W, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-02-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of native and (S)-lysine-bound dihydrodipicolinate synthase from Escherichia coli with improved resolution show new features of biological significance.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2PUR
 
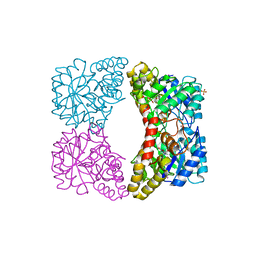 | | Structure of dihydrodipicolinate synthase mutant Thr44Ser at 1.7 A. | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity versus catalytic potency: The role of threonine 44 in Escherichia coli dihydrodipicolinate synthase mediated catalysis.
Biochimie, 91, 2009
|
|
3I7Q
 
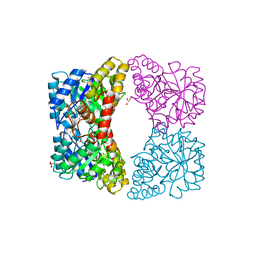 | | Dihydrodipicolinate synthase mutant - K161A | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
3I7R
 
 | | Dihydrodipicolinate synthase - K161R | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
3I7S
 
 | | Dihydrodipicolinate synthase mutant - K161A - with the substrate pyruvate bound in the active site. | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
7QHA
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol | | Descriptor: | Megabody c7HopQ, Putative TRAP-type C4-dicarboxylate transport system, large permease component, ... | | Authors: | North, R.A, Davies, J.S, Morado, D, Dobson, R.C.J. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
8B01
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc | | Descriptor: | DECANE, DOCOSANE, HEXANE, ... | | Authors: | Davies, J.S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2022-09-06 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
8UUB
 
 | | Structure of hypothiocyanous acid reductase (Har) from Streptococcus pneumoniae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Oxidoreductase, ... | | Authors: | Shearer, H.L, Currie, M.J, Dickerhof, N, Dobson, R.C.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hypothiocyanous acid reductase is critical for host colonization and infection by Streptococcus pneumoniae.
J.Biol.Chem., 2024
|
|
8GEK
 
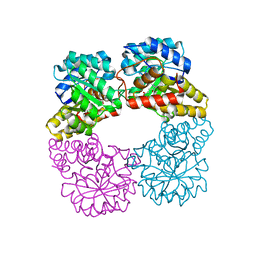 | | Dihydrodipicolinate synthase with pyruvate from Candidatus Liberibacter solanacearum | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase | | Authors: | Gilkes, J.M, Frampton, R.A, Board, A, Sheen, C.R, Smith, G.R, Dobson, R.C.J. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dihydrodipicolinate synthase with pyruvate from the plant pathogen, Candidatus Liberibacter solanacearum
To Be Published
|
|
8THI
 
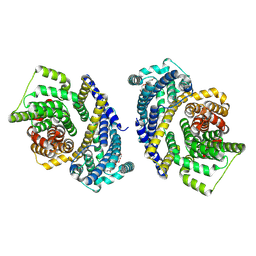 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
8THJ
 
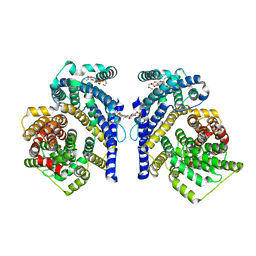 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
7T3E
 
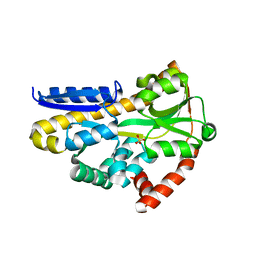 | | Structure of the sialic acid bound Tripartite ATP-independent Periplasmic (TRAP) periplasmic component SiaP from Photobacterium profundum | | Descriptor: | N-acetyl-beta-neuraminic acid, SULFATE ION, TRAP-type C4-dicarboxylate transport system, ... | | Authors: | Davies, J.S, Currie, M.J, North, R.A, Dobson, R.C.J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
6H4E
 
 | | Proteus mirabilis N-acetylneuraminate lyase | | Descriptor: | Putative N-acetylneuraminate lyase, SULFATE ION | | Authors: | North, R.A, Garcia-Bonete, M.J, Goyal, P, Katona, G, Dobson, R.C.J, Friemann, R. | | Deposit date: | 2018-07-21 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | The structure of Proteus mirabilis N-acetylneuraminate lyase reveals an intermolecular disulphide bond
To Be Published
|
|
4EOU
 
 | |
7RUM
 
 | | Endolysin from Escherichia coli O157:H7 phage FTEbC1, LysT84 | | Descriptor: | Endolysin, GLYCEROL | | Authors: | Love, M.J, Billington, C, Dobson, R.C.J. | | Deposit date: | 2021-08-17 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The structure and function of modular Escherichia coli O157:H7 bacteriophage FTBEc1 endolysin, LysT84: defining a new endolysin catalytic subfamily.
Biochem.J., 479, 2022
|
|
3A5F
 
 | |
