8RT0
 
 | | BTV-15 VP5 pH 6.0 | | Descriptor: | 1,2-ETHANEDIOL, Outer capsid protein VP5 | | Authors: | Stuart, D.I, Sutton, G.C. | | Deposit date: | 2024-01-25 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The effect of pH on the structure of Bluetongue virus VP5.
J.Gen.Virol., 105, 2024
|
|
4OTS
 
 | | Crystal Structure of isolated Operophtera brumata CPV18 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stuart, D.I, Sutton, G.C, Axford, D, Ji, X. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-14 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | In cellulo structure determination of a novel cypovirus polyhedrin.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OTV
 
 | | Crystal structure of in cellulo Operophtera brumata CPV18 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stuart, D.I, Sutton, G.C, Axford, D, Ji, X. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In cellulo structure determination of a novel cypovirus polyhedrin.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8B9F
 
 | | Structure of Echovirus 11 complexed with DAF (CD55) calculated from symmetry expansion | | Descriptor: | Complement decay-accelerating factor, Genome polyprotein, SPHINGOSINE | | Authors: | Stuart, D.I, Ren, J, Qin, L, Zhou, D. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
8B8R
 
 | | Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55) | | Descriptor: | DECAY ACCELERATING FACTOR (CD55), SPHINGOSINE, VP1, ... | | Authors: | Stuart, D.I, Ren, J, Zhou, D, Qin, L. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
5FJ5
 
 | | Structure of the in vitro assembled bacteriophage phi6 polymerase complex | | Descriptor: | MAJOR INNER PROTEIN P1 | | Authors: | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | Descriptor: | FAB Heavy chain, FAB Light chain | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3KYJ
 
 | | Crystal structure of the P1 domain of CheA3 in complex with CheY6 from R. sphaeroides | | Descriptor: | CheY6 protein, Putative histidine protein kinase, SODIUM ION | | Authors: | Bell, C.H, Porter, S.L, Armitage, J.P, Stuart, D.I. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex
Plos Biol., 8, 2010
|
|
6J7V
 
 | |
5MUV
 
 | | Atomic structure fitted into a localized reconstruction of bacteriophage phi6 packaging hexamer P4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
1H8T
 
 | | Echovirus 11 | | Descriptor: | 12-AMINO-DODECANOIC ACID, ECHOVIRUS 11 COAT PROTEIN VP1, ECHOVIRUS 11 COAT PROTEIN VP2, ... | | Authors: | Stuart, A, McKee, T, Williams, P.A, Harley, C, Stuart, D.I, Brown, T.D.K, Lea, S.M. | | Deposit date: | 2001-02-15 | | Release date: | 2002-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Determination of the Structure of a Decay Accelerating Factor-Binding Clinical Isolate of Echovirus 11 Allows Mapping of Mutants with Altered Receptor Requirements for Infection
J.Virol., 76, 2002
|
|
7Z3Z
 
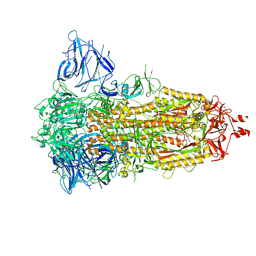 | | Locked Wuhan SARS-CoV2 Prefusion Spike ectodomain with lipid bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, STEARIC ACID, ... | | Authors: | Duyvesteyn, H.M.E, Carrique, L, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2022-03-03 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 Spike harbours a lipid binding pocket which modulates stability of the prefusion trimer
bioRxiv, 2020
|
|
9F9Y
 
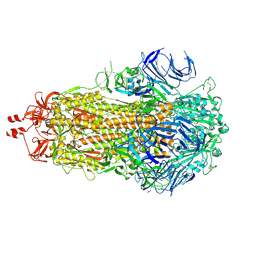 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 2024
|
|
5MQW
 
 | | High-speed fixed-target serial virus crystallography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
6F6I
 
 | | CRYSTAL STRUCTURE OF EBOLAVIRUS GLYCOPROTEIN IN COMPLEX WITH PAROXETINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Envelope glycoprotein, ... | | Authors: | Ren, J, Zhao, Y, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-12-05 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target Identification and Mode of Action of Four Chemically Divergent Drugs against Ebolavirus Infection.
J. Med. Chem., 61, 2018
|
|
6F6N
 
 | | CRYSTAL STRUCTURE OF EBOLAVIRUS GLYCOPROTEIN IN COMPLEX WITH SERTRALINE | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-12-05 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Target Identification and Mode of Action of Four Chemically Divergent Drugs against Ebolavirus Infection.
J. Med. Chem., 61, 2018
|
|
7OOK
 
 | | Bacteriophage PRD1 Major Capsid Protein P3 in complex with CPZ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CHLORIDE ION, ... | | Authors: | Duyvesteyn, H.M.E, Peccati, F, Martinez-Castillo, A, Jimenez-Oses, G, Oksanen, H.M, Stuart, D.I, Abrescia, N.G.A. | | Deposit date: | 2021-05-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Bacteriophage PRD1 as a nanoscaffold for drug loading
Nanoscale, 13, 2021
|
|
1TL1
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW451211 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFINYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1TKT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW426318 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-PROPYLQUINOLIN-2(1H)-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1TKZ
 
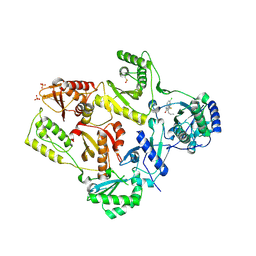 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW429576 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFANYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1TKX
 
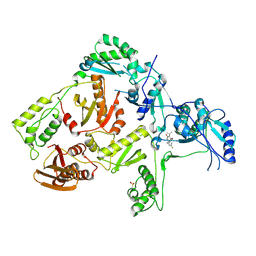 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW490745 | | Descriptor: | 4-[(CYCLOPROPYLETHYNYL)OXY]-6-FLUORO-3-ISOPROPYLQUINOLIN-2(1H)-ONE, Pol polyprotein, Reverse transcriptase, ... | | Authors: | Ren, J, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
J.Med.Chem., 47, 2004
|
|
1TL3
 
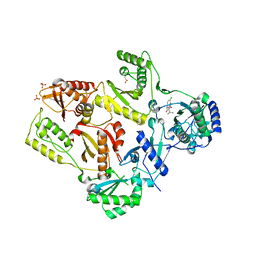 | | Crystal structure of hiv-1 reverse transcriptase in complex with gw450557 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-ISOPROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
7B3Y
 
 | | Structure of a nanoparticle for a COVID-19 vaccine candidate | | Descriptor: | Fibronectin binding protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses.
Nat Commun, 12, 2021
|
|
1QQR
 
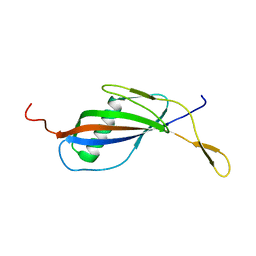 | | CRYSTAL STRUCTURE OF STREPTOKINASE DOMAIN B | | Descriptor: | STREPTOKINASE DOMAIN B | | Authors: | Spraggon, G, Zhang, X.X, Ponting, C.P, Fox, V.F, Phillips, C, Smith, R.A.G, Jones, E.Y, Dobson, C, Stuart, D.I. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Streptokinse Domain B
To be Published
|
|
1QFT
 
 | | HISTAMINE BINDING PROTEIN FROM FEMALE BROWN EAR RHIPICEPHALUS APPENDICULATUS | | Descriptor: | HISTAMINE, PROTEIN (FEMALE-SPECIFIC HISTAMINE BINDING PROTEIN 2) | | Authors: | Paesen, G.C, Adams, P.L, Harlos, K, Nuttal, P.A, Stuart, D.I. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure.
Mol.Cell, 3, 1999
|
|
