2B7A
 
 | | The structural basis of Janus Kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Tyrosine-protein kinase JAK2 | | Authors: | Lucet, I.S, Fantino, E, Styles, M, Bamert, R, Patel, O, Broughton, S.E, Walter, M, Burns, C.J, Treutlein, H, Wilks, A.F, Rossjohn, J. | | Deposit date: | 2005-10-04 | | Release date: | 2006-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor.
Blood, 107, 2006
|
|
7SHW
 
 | |
7N3V
 
 | | Crystal structure of Mycobacterium smegmatis LmcA | | Descriptor: | GLYCEROL, LmcA, SULFATE ION | | Authors: | Patel, O, Lucet, I, Panjikar, S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the putative cell-wall lipoglycan biosynthesis protein LmcA from Mycobacterium smegmatis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5JZJ
 
 | | Crystal structure of DCLK1-KD in complex with AMPPN | | Descriptor: | AMP PHOSPHORAMIDATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1.
Structure, 24, 2016
|
|
5VE6
 
 | | Crystal structure of Sugen kinase 223 | | Descriptor: | Tyrosine-protein kinase SgK223 | | Authors: | Patel, O, Lucet, I, Panjikar, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association.
Nat Commun, 8, 2017
|
|
5JZN
 
 | | Crystal structure of DCLK1-KD in complex with NVP-TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, SULFATE ION, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1.
Structure, 24, 2016
|
|
8DGP
 
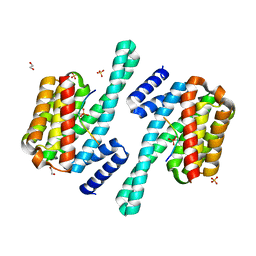 | | 14-3-3 epsilon bound to phosphorylated PEAK3 (pS69) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Phosphorylated PEAK3 (pS69) peptide, ... | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
2OGV
 
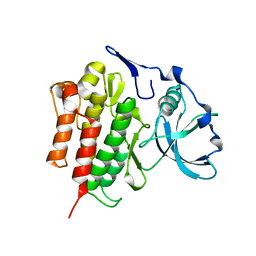 | | Crystal Structure of the Autoinhibited Human c-Fms Kinase Domain | | Descriptor: | Macrophage colony-stimulating factor 1 receptor precursor | | Authors: | Walter, M, Lucet, I.S, Patel, O, Broughton, S.E, Bamert, R, Williams, N.K, Fantino, E, Wilks, A.F, Rossjohn, J. | | Deposit date: | 2007-01-09 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The 2.7 A crystal structure of the autoinhibited human c-Fms kinase domain.
J.Mol.Biol., 367, 2007
|
|
4QRP
 
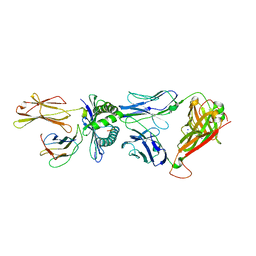 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL and DD31 TCR | | Descriptor: | Beta-2-microglobulin, DD31 TCR alpha chain, DD31 TCR beta chain, ... | | Authors: | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
8DGO
 
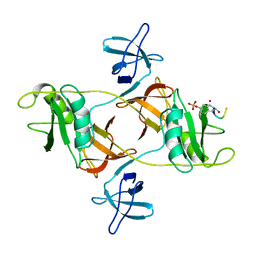 | |
8DGN
 
 | |
8DGM
 
 | | 14-3-3 epsilon bound to phosphorylated PEAK1 (pT1165) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Inactive tyrosine-protein kinase PEAK1 | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
7KX6
 
 | | Crystal structure of DCLK1-KD in complex with XMD8-85 | | Descriptor: | 2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-03 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
4QRQ
 
 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
7KXW
 
 | | Crystal structure of DCLK1-KD in complex with DCLK1-IN-1 | | Descriptor: | 2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-11-methyl-5-(2,2,2-trifluoroethyl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase DCLK1, ... | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
7KX8
 
 | | Crystal structure of DCLK1-Cter in complex with FMF-03-055-1 | | Descriptor: | 5-ethyl-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-11-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-03 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
6C7Y
 
 | | Crystal structure of inhibitory protein SOCS1 in complex with JAK1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Liau, N.P.D, Laktyushin, A, Lucet, I.S, Murphy, J.M, Yao, S, Callaghan, K, Nicola, N.A, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | The molecular basis of JAK/STAT inhibition by SOCS1.
Nat Commun, 9, 2018
|
|
3FUP
 
 | | Crystal structures of JAK1 and JAK2 inhibitor complexes | | Descriptor: | 3-{(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}-3-oxopropanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Williams, N.K, Bamert, R.S, Patel, O, Fantino, E, Rossjohn, J, Lucet, I.S. | | Deposit date: | 2009-01-14 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains.
J.Mol.Biol., 387, 2009
|
|
