6M4O
 
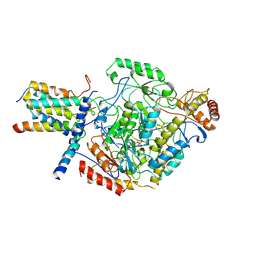 | | Cryo-EM structure of the monomeric SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-03-08 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6M4N
 
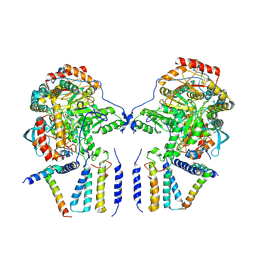 | | Cryo-EM structure of the dimeric SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-03-07 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7CQI
 
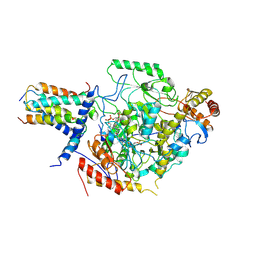 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7CQK
 
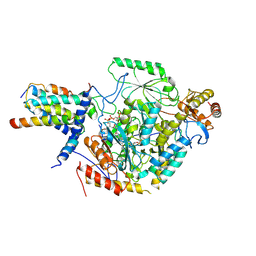 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4F5A
 
 | | Triple mutant Src SH2 domain bound to phosphate ion | | Descriptor: | PHOSPHATE ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F59
 
 | | Triple mutant Src SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F5B
 
 | | Triple mutant Src SH2 domain bound to phosphotyrosine | | Descriptor: | O-PHOSPHOTYROSINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
3MAZ
 
 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
1H3H
 
 | | Structural Basis for Specific Recognition of an RxxK-containing SLP-76 peptide by the Gads C-terminal SH3 domain | | Descriptor: | GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Liu, Q, Berry, D, Nash, P, Pawson, T, McGlade, C.J, Li, S.S. | | Deposit date: | 2002-09-03 | | Release date: | 2003-03-06 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Specific Binding of the Gads SH3 Domain to an Rxxk Motif-Containing Slp-76 Peptide: A Novel Mode of Peptide Recognition
Mol.Cell, 11, 2003
|
|
4GXB
 
 | | Structure of the SNX17 atypical FERM domain bound to the NPxY motif of P-selectin | | Descriptor: | GLYCEROL, P-selectin, Sorting nexin-17 | | Authors: | Ghai, R, Bugarcic, A, Liu, H, Norwood, S.J, Li, S.S, Teasdale, R.D, Collins, B.M. | | Deposit date: | 2012-09-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6IV5
 
 | | Crystal structure of arabidopsis N6-mAMP deaminase MAPDA | | Descriptor: | Adenosine/AMP deaminase family protein, PHOSPHATE ION, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6J23
 
 | | Crystal structure of arabidopsis ADAL complexed with GMP | | Descriptor: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6J4T
 
 | | Crystal structure of arabidopsis ADAL complexed with IMP | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6E8M
 
 | |
6E8H
 
 | | Legionella Longbeachae LeSH (Llo2327) | | Descriptor: | CHLORIDE ION, LeSH (Llo2327) | | Authors: | Kaneko, T, Li, S.S.C. | | Deposit date: | 2018-07-29 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification and characterization of a large family of superbinding bacterial SH2 domains.
Nat Commun, 9, 2018
|
|
6E8K
 
 | |
6E8I
 
 | |
2NNZ
 
 | | Solution structure of the hypothetical protein AF2241 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein | | Authors: | Ai, X, Semesib, A, Yee, A, Arrowsmith, C.H, Li, S.S.C, Choy, W.Y, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2006-10-24 | | Release date: | 2007-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Hypothetical protein AF2241 from Archaeoglobus fulgidus adopts a cyclophilin-like fold.
J.Biomol.Nmr, 38, 2007
|
|
1WM3
 
 | |
1WM2
 
 | |
2AWT
 
 | | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2) | | Descriptor: | Small ubiquitin-related modifier 2 | | Authors: | Chang, C.K, Wang, Y.H, Chung, T.L, Chang, C.F, Li, S.S.L, Huang, T.H. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2)
To be Published
|
|
2JMB
 
 | | Solution structure of the protein Atu4866 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu4866 | | Authors: | Ai, X, Semesi, A, Yee, A, Arrowsmith, C.H, Li, S.S.C, Choy, W, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2006-11-01 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein Atu4866 from Agrobacterium tumefaciens
To be Published
|
|
2JPI
 
 | | NMR structure of PA4090 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical protein | | Authors: | Ai, X, Semesi, A, Yee, A, Arrowsmith, C.H, Li, S.S.C, Choy, W, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-05-13 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | chemical shift assignments of PA4090 from Pseudomonas aeruginosa
To be Published
|
|
