8DUW
 
 | | HnRNPA2 D290V LCD PM2 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
8DU2
 
 | | HnRNPA2 D290V LCD PM1 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
8EC7
 
 | | HnRNPA2 D290V LCD PM3 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
6UFR
 
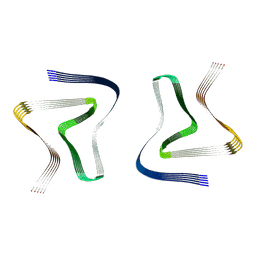 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NK4
 
 | |
2GLS
 
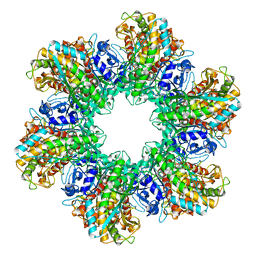 | |
6ODG
 
 | | SVQIVY, Crystal Structure of a tau protein fragment | | Descriptor: | Microtubule-associated protein tau | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Seidler, P.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure-based inhibitors halt prion-like seeding by Alzheimer's disease-and tauopathy-derived brain tissue samples.
J.Biol.Chem., 294, 2019
|
|
2MLT
 
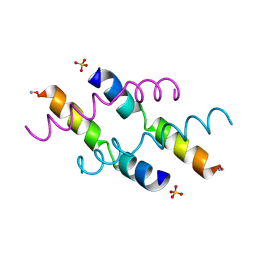 | |
6N4P
 
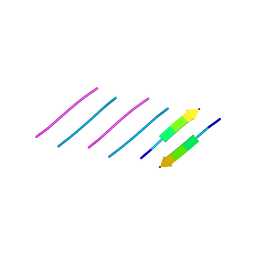 | |
1BYZ
 
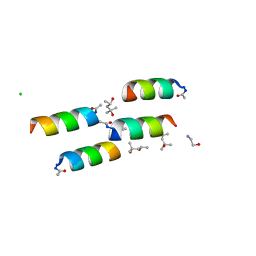 | | DESIGNED PEPTIDE ALPHA-1, P1 FORM | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Prive, G.G, Anderson, D.H, Wesson, L, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-20 | | Release date: | 1998-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Packed protein bilayers in the 0.90 A resolution structure of a designed alpha helical bundle.
Protein Sci., 8, 1999
|
|
7N8R
 
 | | FGTGFG segment from the Nucleoporin p54, residues 63-68 | | Descriptor: | FGTGFG segment from the Nucleoporin p54, residues 63-68, trifluoroacetic acid | | Authors: | Hughes, M.P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Extended beta-Strands Contribute to Reversible Amyloid Formation.
Acs Nano, 16, 2022
|
|
1A2W
 
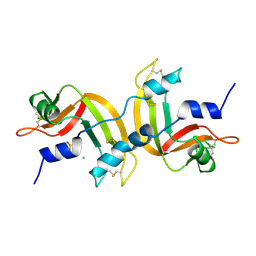 | | CRYSTAL STRUCTURE OF A 3D DOMAIN-SWAPPED DIMER OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A, SULFATE ION | | Authors: | Liu, Y, Hart, P.J, Schlunegger, M.P, Eisenberg, D.S. | | Deposit date: | 1998-01-12 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a 3D domain-swapped dimer of RNase A at a 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1B4T
 
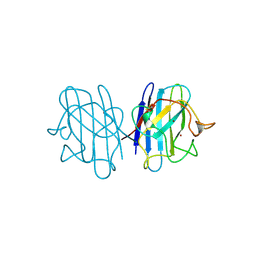 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1B4L
 
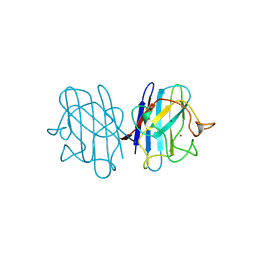 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1AL1
 
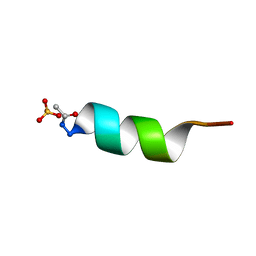 | | CRYSTAL STRUCTURE OF ALPHA1: IMPLICATIONS FOR PROTEIN DESIGN | | Descriptor: | ALPHA HELIX PEPTIDE: ELLKKLLEELKG, SULFATE ION | | Authors: | Hill, C.P, Anderson, D.H, Wesson, L, Degrado, W.F, Eisenberg, D. | | Deposit date: | 1990-07-02 | | Release date: | 1991-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of alpha 1: implications for protein design.
Science, 249, 1990
|
|
1BGC
 
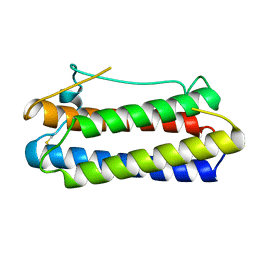 | |
1BGE
 
 | |
1BP1
 
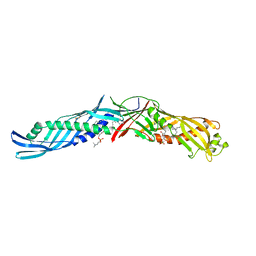 | | CRYSTAL STRUCTURE OF BPI, THE HUMAN BACTERICIDAL PERMEABILITY-INCREASING PROTEIN | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Beamer, L.J, Carroll, S.F, Eisenberg, D. | | Deposit date: | 1997-04-08 | | Release date: | 1997-09-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution.
Science, 276, 1997
|
|
1BGD
 
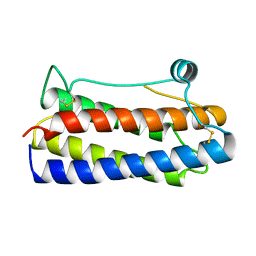 | |
1AZV
 
 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | Descriptor: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1997-11-21 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
8DDF
 
 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
8TEQ
 
 | | Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) G269V mutant, amyloid fiber | | Descriptor: | TRK-fused gene protein Low Complexity Domain G269V mutant | | Authors: | Rosenberg, G.M, Sawaya, M.R, Boyer, D.R, Ge, P, Abskharon, R, Eisenberg, D.S. | | Deposit date: | 2023-07-06 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Fibril structures of TFG protein mutants validate the identification of TFG as a disease-related amyloid protein by the IMPAcT method.
Pnas Nexus, 2, 2023
|
|
8TER
 
 | | Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) P285L mutant, amyloid fiber | | Descriptor: | TRK-fused gene protein Low Complexity Domain P285L mutant | | Authors: | Rosenberg, G.M, Sawaya, M.R, Boyer, D.R, Ge, P, Abskharon, R, Eisenberg, D.S. | | Deposit date: | 2023-07-06 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Fibril structures of TFG protein mutants validate the identification of TFG as a disease-related amyloid protein by the IMPAcT method.
Pnas Nexus, 2, 2023
|
|
8FQ7
 
 | | Nanobody with WIW inserted in CDR3 loop to Inhibit Growth of Alzheimer's Tau fibrils | | Descriptor: | GLYCEROL, Nanobody with WIW insert in CDR3 loop to target tau fibrils | | Authors: | Abskharon, R, Sawaya, M.R, Cascio, D.C, Eisenberg, D.S. | | Deposit date: | 2023-01-05 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of nanobodies that inhibit seeding of Alzheimer's patient-extracted tau fibrils.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DDG
 
 | |
