1Q5R
 
 | | The Rhodococcus 20S proteasome with unprocessed pro-peptides | | Descriptor: | proteasome alpha-type subunit 1, proteasome beta-type subunit 1 | | Authors: | Kwon, Y.D, Nagy, I, Adams, P.D, Baumeister, W, Jap, B.K. | | Deposit date: | 2003-08-08 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly.
J.Mol.Biol., 335, 2004
|
|
1Q5Q
 
 | | The Rhodococcus 20S proteasome | | Descriptor: | proteasome alpha-type subunit 1, proteasome beta-type subunit 1 | | Authors: | Kwon, Y.D, Nagy, I, Adams, P.D, Baumeister, W, Jap, B.K. | | Deposit date: | 2003-08-08 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly.
J.Mol.Biol., 335, 2004
|
|
5WDF
 
 | |
3TGR
 
 | | Crystal structure of unliganded HIV-1 clade C strain C1086 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGQ
 
 | | Crystal structure of unliganded HIV-1 clade B strain YU2 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 YU2 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGS
 
 | | Crystal structure of HIV-1 clade C strain C1086 gp120 core in complex with NBD-556 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 core, N-(4-chlorophenyl)-N'-(2,2,6,6-tetramethylpiperidin-4-yl)ethanediamide | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TIH
 
 | |
4DVV
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with AS-I-261 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-{[(4S,5S)-5-(aminomethyl)-2,2-dimethyl-1,3-dioxolan-4-yl]methyl}-N'-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2012-02-23 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
4DVS
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with NBD-557 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(4-bromophenyl)-N'-(2,2,6,6-tetramethylpiperidin-4-yl)ethanediamide, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2012-02-23 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
4DVT
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with AS-II-37 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(4-chloro-3-fluorophenyl)-N'-[(1S)-1,2,3,4-tetrahydroisoquinolin-1-ylmethyl]ethanediamide, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2012-02-23 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
4DVX
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with MAE-II-188 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(4-chloro-3-fluorophenyl)-N'-{[(3R)-1-cyclopropylpyrrolidin-3-yl]methyl}ethanediamide, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2012-02-23 | | Release date: | 2013-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
4DVW
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with MAE-II-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(4-chloro-3-fluorophenyl)-N'-[(3aS,6aS)-hexahydrocyclopenta[c]pyrrol-3a(1H)-ylmethyl]ethanediamide, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2012-02-23 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
4DVR
 
 | |
8DW2
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW3
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8FA0
 
 | |
8F9Z
 
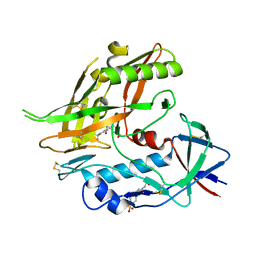 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with NBD-14204, an HIV-1 gp120 antagonist | | Descriptor: | (5M)-N-{(1S)-2-amino-1-[5-(hydroxymethyl)-1,3-thiazol-2-yl]ethyl}-5-[5-(trifluoromethyl)pyridin-2-yl]-1H-pyrrole-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Antiviral Activity and Crystal Structures of HIV-1 gp120 Antagonists.
Int J Mol Sci, 23, 2022
|
|
2H6J
 
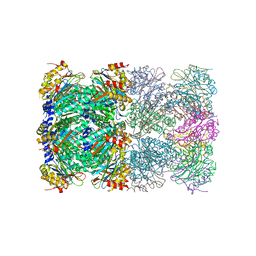 | | Crystal Structure of the Beta F145A Rhodococcus Proteasome | | Descriptor: | Proteasome alpha-type subunit 1, Proteasome beta-type subunit 1 | | Authors: | Kwon, Y.D. | | Deposit date: | 2006-05-31 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Proteasome assembly triggers a switch required for active-site maturation.
Structure, 14, 2006
|
|
7TZ0
 
 | |
7TYZ
 
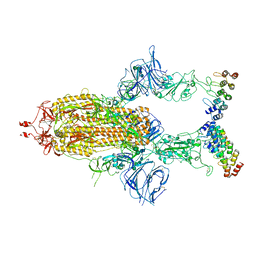 | | Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DARPin FSR22, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
4JB9
 
 | | Crystal structure of antibody VRC06 in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, antibody VRC06 heavy chain, antibody VRC06 light chain, ... | | Authors: | Kwon, Y.D, Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-19 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization.
Science, 340, 2013
|
|
5U6E
 
 | | Crystal structure of clade A/E HIV-1 gp120 core in complex with NBD-14010 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-{(1S)-2-amino-1-[5-(hydroxymethyl)-4-methyl-1,3-thiazol-2-yl]ethyl}-5-(4-chloro-3-fluorophenyl)-1H-pyrrole-2-carboxamide, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis, Antiviral Potency, in Vitro ADMET, and X-ray Structure of Potent CD4 Mimics as Entry Inhibitors That Target the Phe43 Cavity of HIV-1 gp120.
J. Med. Chem., 60, 2017
|
|
5IQ7
 
 | | Crystal structure of 10E8-S74W Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8-S74W Heavy Chain, 10E8-S74W Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2869 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
5IQ9
 
 | | Crystal structure of 10E8v4 Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8v4 Heavy Chain, 10E8v4 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
