6YQT
 
 | | CRYSTAL STRUCTURE OF HUMAN CA II IN COMPLEX WITH A SULFONAMIDE DERIVATIVE OF 2-MERCAPTOBENZOXAZOLE | | Descriptor: | 2-sulfanylidene-3~{H}-1,3-benzoxazole-5-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2020-04-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 2-Mercaptobenzoxazoles: a class of carbonic anhydrase inhibitors with a novel binding mode to the enzyme active site.
Chem.Commun.(Camb.), 56, 2020
|
|
2JVE
 
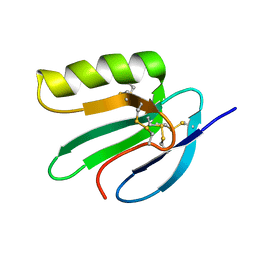 | | Solution structure of the extracellular domain of Prod1, a protein implicated in proximodistal identity during amphibian limb regeneration | | Descriptor: | Prod 1 | | Authors: | Garza-Garcia, A, Harris, R, Esposito, D, Driscoll, P.C. | | Deposit date: | 2007-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phylogenetics of Prod1, a member of the three-finger protein superfamily implicated in salamander limb regeneration.
Plos One, 4, 2009
|
|
2JP0
 
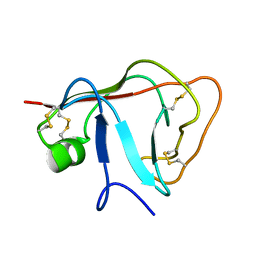 | | Solution structure of the N-terminal extraceullular domain of the lymphocyte receptor CD5 calculated using inferential structure determination (ISD) | | Descriptor: | T-cell surface glycoprotein CD5 | | Authors: | Garza-Garcia, A, Harris, R, Esposito, D, Driscoll, P.C, Rieping, W. | | Deposit date: | 2007-04-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and conformational plasticity of the N-terminal scavenger receptor cysteine-rich domain of human CD5
J.Mol.Biol., 378, 2008
|
|
2JOP
 
 | |
6SVF
 
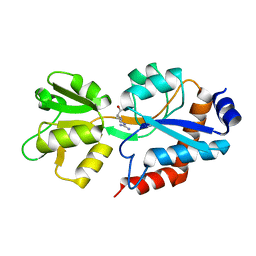 | | Crystal structure of the P235GK mutant of ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Vitagliano, L, Berisio, R, Esposito, L, Balasco, N, Smaldone, G, Ruggiero, A. | | Deposit date: | 2019-09-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The non-swapped monomeric structure of the arginine-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6P0Z
 
 | | Crystal structure of N-acetylated KRAS (2-169) bound to GDP and Mg | | Descriptor: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, GTPase KRas, ... | | Authors: | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.011 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
6MBQ
 
 | | Crystal structure of Mg-free wild-type KRAS (2-166) bound to GMPPNP in the state 1 conformation | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Dharmaiah, S, Davies, D.R, Abendroth, J, Gillette, W.G, Stephen, A.G, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
4EY0
 
 | | Structure of tandem SH2 domains from PLCgamma1 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
4FBN
 
 | | Insights into structural integration of the PLCgamma regulatory region and mechanism of autoinhibition and activation based on key roles of SH2 domains | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
2C5L
 
 | | Structure of PLC epsilon Ras association domain with hRas | | Descriptor: | GLYCEROL, GTPASE HRAS, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roe, S.M, Bunney, T.D, Katan, M, Pearl, L.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon
Mol.Cell, 21, 2006
|
|
6MBU
 
 | | Crystal structure of wild-type KRAS (1-169) bound to GDP and Mg (Space group P3) | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
6MBT
 
 | | Crystal structure of wild-type KRAS bound to GDP and Mg (Space group C2) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
6M9W
 
 | | Structure of Mg-free KRAS4b (2-169) bound to GDP with the switch-I in fully open conformation | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
5TAR
 
 | | Crystal structure of farnesylated and methylated kras4b in complex with PDE-delta (crystal form II - with ordered hypervariable region) | | Descriptor: | 1,2-ETHANEDIOL, FARNESYL, GTPase KRas, ... | | Authors: | Dharmaiah, S, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2016-09-10 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of recognition of farnesylated and methylated KRAS4b by PDE delta.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5TB5
 
 | | Crystal structure of full-length farnesylated and methylated KRAS4b in complex with PDE-delta (crystal form I - with partially disordered hypervariable region) | | Descriptor: | 1,2-ETHANEDIOL, FARNESYL, GTPase KRas, ... | | Authors: | Dharmaiah, S, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2016-09-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of recognition of farnesylated and methylated KRAS4b by PDE delta.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8U1N
 
 | |
8U1M
 
 | |
8T71
 
 | | Crystal Structure of WT KRAS4a with bound GDP and Mg ion | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
8T72
 
 | | Crystal structure of WT KRAS4a with bound GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
8T75
 
 | |
8T73
 
 | | Crystal structure of KRAS4a-R151G with bound GDP and Mg ion | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
8T74
 
 | |
8U1L
 
 | |
7BEZ
 
 | |
7BF0
 
 | | Zn-bound domain 3 of CDCA1 in complex with carbon dioxide | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, Cadmium-specific carbonic anhydrase, ... | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2020-12-30 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Zeta-carbonic anhydrases show CS 2 hydrolase activity: A new metabolic carbon acquisition pathway in diatoms?
Comput Struct Biotechnol J, 19, 2021
|
|
