3BI6
 
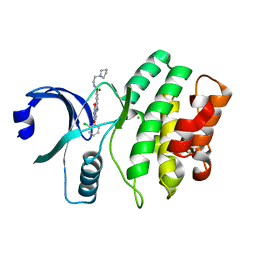 | | Wee1 kinase complex with inhibitor PD352396 | | Descriptor: | 4-(2-chlorophenyl)-9-hydroxy-6-methyl-1,3-dioxo-N-(2-pyrrolidin-1-ylethyl)pyrrolo[3,4-g]carbazole-8-carboxamide, CHLORIDE ION, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of soluble 8-substituted 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of the Wee1 and Chk1 checkpoint kinases.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BIZ
 
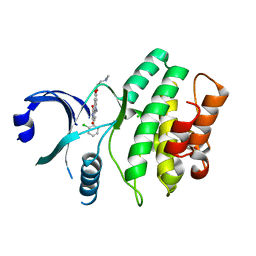 | | Wee1 kinase complex with inhibitor PD331618 | | Descriptor: | 4-(2-chlorophenyl)-8-[3-(dimethylamino)propoxy]-9-hydroxy-6-methylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-dione, CHLORIDE ION, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2007-12-02 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of soluble 8-substituted 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of the Wee1 and Chk1 checkpoint kinases.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4GDM
 
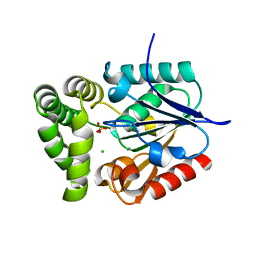 | | Crystal Structure of E.coli MenH | | Descriptor: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | Deposit date: | 2012-07-31 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
4GKM
 
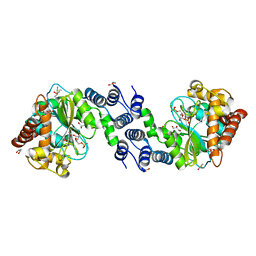 | | Bianthranilate-like analogue bound in the outer site of anthranilate phosphoribosyltransferase (AnPRT; trpD) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-[(2-carboxyphenyl)amino]-5-methylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Evans, G.L, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-08-13 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6683 Å) | | Cite: | Repurposing the Chemical Scaffold of the Anti-Arthritic Drug Lobenzarit to Target Tryptophan Biosynthesis in Mycobacterium tuberculosis.
Chembiochem, 15, 2014
|
|
4GEG
 
 | | Crystal Structure of E.coli MenH Y85F Mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, ... | | Authors: | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
4GIU
 
 | | Bianthranilate-like analogue bound in inner site of anthranilate phosphoribosyltransferase (AnPRT; trpD). | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-[(2-carboxy-5-methylphenyl)amino]-3-methylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Evans, G.L, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-08-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Repurposing the Chemical Scaffold of the Anti-Arthritic Drug Lobenzarit to Target Tryptophan Biosynthesis in Mycobacterium tuberculosis.
Chembiochem, 15, 2014
|
|
3C8N
 
 | |
3CQE
 
 | | Wee1 kinase complex with inhibitor PD074291 | | Descriptor: | 8-bromo-4-(2-chlorophenyl)-N-(2-hydroxyethyl)-6-methyl-1,3-dioxo-1,2,3,6-tetrahydropyrrolo[3,4-e]indole-7-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Squire, C.J, Baker, E.N. | | Deposit date: | 2008-04-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Determinants of Wee1 Inhibitor Selectivity
To be Published
|
|
3CR0
 
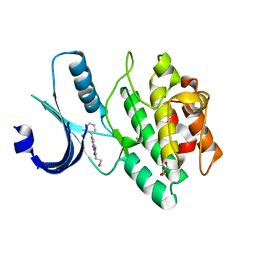 | | Wee1 kinase complex with inhibitor PD259_809 | | Descriptor: | 4-(2-chlorophenyl)-8-(2-hydroxyethyl)-6-methylpyrrolo[3,4-e]indole-1,3(2H,6H)-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Squire, C.J, Baker, E.N. | | Deposit date: | 2008-04-03 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of Wee1 inhibitor selectivity
To be Published
|
|
3CEV
 
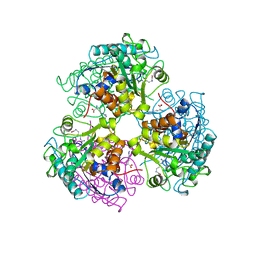 | | ARGINASE FROM BACILLUS CALDEVELOX, COMPLEXED WITH L-ARGININE | | Descriptor: | ARGININE, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
4HSS
 
 | |
4HSQ
 
 | |
3DA8
 
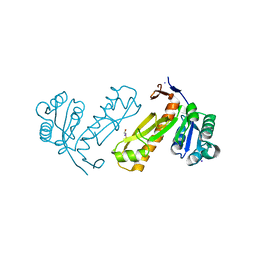 | | Crystal structure of PurN from Mycobacterium tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-05-28 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
3DCJ
 
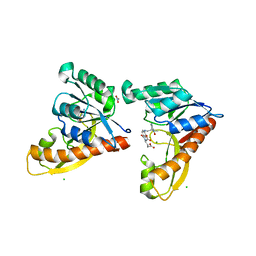 | | Crystal structure of glycinamide formyltransferase (PurN) from Mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-tetrahydrofolic acid derivative | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-06-03 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
3D98
 
 | |
3D8V
 
 | |
4IDS
 
 | |
4IJ1
 
 | | Bianthranilate-like analogue bound to anthranilate phosphoribosyltransferase (AnPRT; trpD) in absence of substrates. | | Descriptor: | 2,2'-iminodibenzoic acid, Anthranilate phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Evans, G.L, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-12-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Repurposing the Chemical Scaffold of the Anti-Arthritic Drug Lobenzarit to Target Tryptophan Biosynthesis in Mycobacterium tuberculosis.
Chembiochem, 15, 2014
|
|
4IDM
 
 | |
4IHI
 
 | |
3E8W
 
 | | Crystal Structure of Epiphyas postvittana Takeout 1 | | Descriptor: | Takeout-like protein 1, Ubiquinone-8 | | Authors: | Hamiaux, C, Stanley, D, Greenwood, D.R, Baker, E.N, Newcomb, R.D. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Epiphyas postvittana takeout 1 with bound ubiquinone supports a role as ligand carriers for takeout proteins in insects
J.Biol.Chem., 284, 2009
|
|
3E3A
 
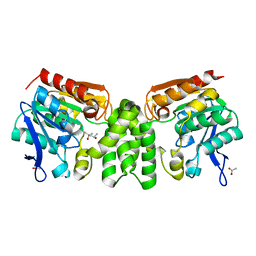 | | The Structure of Rv0554 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, POSSIBLE PEROXIDASE BPOC | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2008-08-06 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3E8T
 
 | | Crystal Structure of Epiphyas postvittana Takeout 1 | | Descriptor: | Takeout-like protein 1, Ubiquinone-8 | | Authors: | Hamiaux, C, Stanley, D, Greenwood, D.R, Baker, E.N, Newcomb, R.D. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of Epiphyas postvittana takeout 1 with bound ubiquinone supports a role as ligand carriers for takeout proteins in insects
J.Biol.Chem., 284, 2009
|
|
3F8L
 
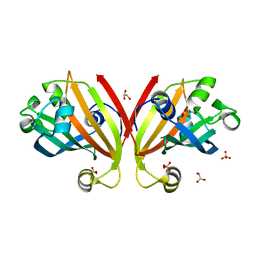 | | Crystal Structure of the Effector Domain of PhnF from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, HTH-type transcriptional repressor phnF, SULFATE ION | | Authors: | Busby, J.N, Gebhard, S, Cook, G.M, Baker, E.N, Lott, S.J, Money, V.A. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of PhnF, a GntR-family transcription regulator in Mycobacterium smegmatis
To be Published
|
|
4JDC
 
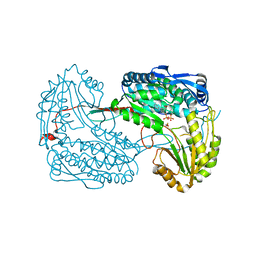 | |
