2O6F
 
 | | Structure of metal- free rTp34 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2O6D
 
 | | Structure of native rTp34 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2O6E
 
 | | Structure of native rTp34 from Treponema pallidum from zinc-soaked crystals | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2O6C
 
 | | Structure of selenomethionyl rTp34 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
3KY9
 
 | | Autoinhibited Vav1 | | Descriptor: | Proto-oncogene vav, ZINC ION | | Authors: | Tomchick, D.R, Rosen, M.K, Machius, M, Yu, B. | | Deposit date: | 2009-12-04 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Structural and Energetic Mechanisms of Cooperative Autoinhibition and Activation of Vav1
Cell(Cambridge,Mass.), 140, 2010
|
|
3TCP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
2F5Z
 
 | | Crystal Structure of Human Dihydrolipoamide Dehydrogenase (E3) Complexed to the E3-Binding Domain of Human E3-Binding Protein | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, Pyruvate dehydrogenase protein X component, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Insight into Interactions between Dihydrolipoamide Dehydrogenase (E3) and E3 Binding Protein of Human Pyruvate Dehydrogenase Complex.
Structure, 14, 2006
|
|
2FQX
 
 | | PnrA from Treponema pallidum complexed with guanosine | | Descriptor: | GUANOSINE, Membrane lipoprotein tmpC | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum
J.Biol.Chem., 281, 2006
|
|
3U3B
 
 | |
5V5V
 
 | | Complex of NLGN2 with MDGA1 Ig1-Ig2 | | Descriptor: | MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-2 | | Authors: | Gangwar, S.P, Machius, M, Rudenko, G. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Molecular Mechanism of MDGA1: Regulation of Neuroligin 2:Neurexin Trans-synaptic Bridges.
Neuron, 94, 2017
|
|
5VPB
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-I crystal | | Descriptor: | CHLORIDE ION, Protein fosB, Transcription factor jun-D | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPD
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-III crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
6NYT
 
 | | Munc13-1 C2B-domain, calcium bound | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tomchick, D.R, Rizo, J, Machius, M, Lu, J. | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis.
Nat. Struct. Mol. Biol., 17, 2010
|
|
6NYC
 
 | | Munc13-1 C2B-domain, calcium free | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Munc13-1 | | Authors: | Tomchick, D.R, Rizo, J, Machius, M, Lu, J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis.
Nat. Struct. Mol. Biol., 17, 2010
|
|
2J9F
 
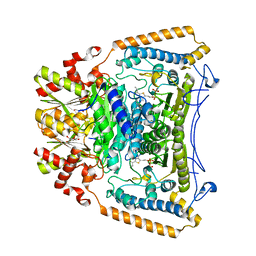 | | Human branched-chain alpha-ketoacid dehydrogenase-decarboxylase E1b | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Jun, L, Machius, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2006-11-07 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Two Active Sites in Human Branched-Chain Alpha- Keto Acid Dehydrogenase Operate Independently without an Obligatory Alternating-Site Mechanism.
J.Biol.Chem., 282, 2007
|
|
5T09
 
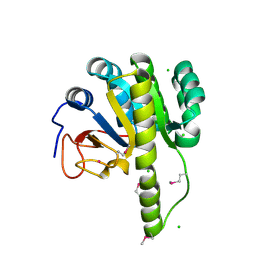 | | The structure of the type III effector HopBA1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Type III secretion system effector HopBA1 | | Authors: | Cherkis, K, Machius, M, Nishimura, M.T, Dangl, J.L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2UXN
 
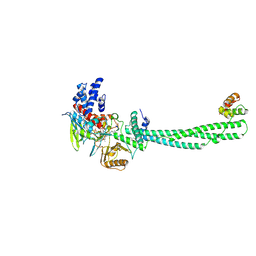 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | Descriptor: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2UXX
 
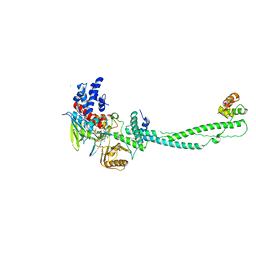 | | Human LSD1 Histone Demethylase-CoREST in complex with an FAD- tranylcypromine adduct | | Descriptor: | CHLORIDE ION, FAD-trans-2-Phenylcyclopropylamine Adduct, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-30 | | Release date: | 2007-08-21 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for the Inhibition of the Lsd1 Histone Demethylase by the Antidepressant Trans-2-Phenylcyclopropylamine.
Biochemistry, 46, 2007
|
|
1U5B
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, GLYCEROL, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-07-27 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
5K0K
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2434 | | Descriptor: | 15-{4-[(4-methylpiperazin-1-yl)methyl]phenyl}-4,5,6,7,9,10,11,12-octahydro-2,16-(azenometheno)pyrrolo[2,1-d][1,3,5,9]te traazacyclotetradecin-8(3H)-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, X, Liu, J, Zhang, W, Stashko, M.A, Nichols, J, DeRyckere, D, Miley, M.J, Norris-Drouin, J, Chen, Z, Machius, M, Wood, E, Graham, D.K, Earp, H.S, Graham, K, Kireev, D, Frye, S.V. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Design and Synthesis of Novel Macrocyclic Mer Tyrosine Kinase Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5K0X
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
4M3Q
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1917 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Zhang, W, Zhang, D, Stashko, M.A, DeRyckere, D, Hunter, D, Kireev, D.B, Miley, M, Cummings, C, Lee, M, Norris-Drouin, J, Stewart, W.M, Sather, S, Zhou, Y, Kirkpatrick, G, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4GOU
 
 | | Crystal structure of an RGS-RhoGEF from Entamoeba histolytica | | Descriptor: | EhRGS-RhoGEF | | Authors: | Bosch, D.E, Kimple, A.J, Muller, R.E, Willard, F.S, Machius, M, Siderovski, D.P. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-09 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants of RGS-RhoGEF Signaling Critical to Entamoeba histolytica Pathogenesis.
Structure, 21, 2013
|
|
4FID
 
 | | Crystal structure of a heterotrimeric G-Protein subunit from entamoeba histolytica, EHG-ALPHA-1 | | Descriptor: | G protein alpha subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Bosch, D.E, Kimple, A.J, Muller, R.E, Gigure, P.M, Willard, F.S, Machius, M, Temple, B.R, Siderovski, D.P. | | Deposit date: | 2012-06-08 | | Release date: | 2012-11-28 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Heterotrimeric G-protein Signaling Is Critical to Pathogenic Processes in Entamoeba histolytica.
Plos Pathog., 8, 2012
|
|
4MHA
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1817 | | Descriptor: | 2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-(4-sulfamoylbenzyl)pyrimidine-5-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, W, Mciver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, D.B.M, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, Dipaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
