3K7V
 
 | | Protein phosphatase 2A core complex bound to dinophysistoxin-1 | | Descriptor: | (2R)-3-[(2S,5R,6R,8S)-8-{(1R,2E)-3-[(2R,4a'R,5R,6'S,8'R,8a'S)-6'-{(1S,3S)-3-[(2S,3R,6R,11R)-3,11-dimethyl-1,7-dioxaspiro[5.5]undec-2-yl]-1-hydroxybutyl}-8'-hydroxy-7'-methylideneoctahydro-3H,3'H-spiro[furan-2,2'-pyrano[3,2-b]pyran]-5-yl]-1-methylprop-2-en-1-yl}-5-hydroxy-10-methyl-1,7-dioxaspiro[5.5]undec-10-en-2-yl]-2-hydroxy-2-methylpropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Jeffrey, P.D, Huhn, J, Shi, Y. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A structural basis for the reduced toxicity of dinophysistoxin-2.
Chem.Res.Toxicol., 22, 2009
|
|
7KGX
 
 | | Structure of PQS Response Protein PqsE in Complex with 4-(3-(2-methyl-2-morpholinobutyl)ureido)-N-(thiazol-2-yl)benzamide | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-({[(2R)-2-methyl-2-(morpholin-4-yl)butyl]carbamoyl}amino)-N-(1,3-thiazol-2-yl)benzamide, FE (III) ION | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor Mimetic Mutations in the Pseudomonas aeruginosa PqsE Enzyme Reveal a Protein-Protein Interaction with the Quorum-Sensing Receptor RhlR That Is Vital for Virulence Factor Production.
Acs Chem.Biol., 16, 2021
|
|
7KGW
 
 | |
1DKK
 
 | | BOBWHITE QUAIL LYSOZYME WITH NITRATE | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
1DKJ
 
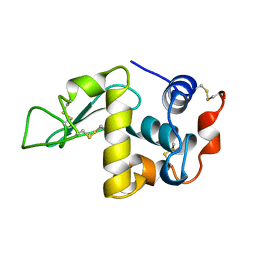 | | BOBWHITE QUAIL LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
7S97
 
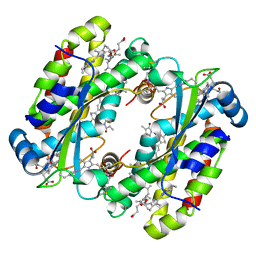 | | Structure of the Photoacclimated Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7S96
 
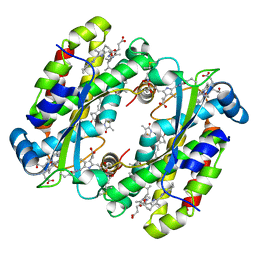 | | Structure of the Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TLF
 
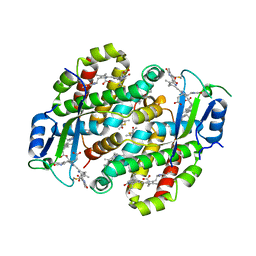 | | Structure of the photoacclimated Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOERYTHROBILIN, Phycoerythrin alpha-subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TJA
 
 | | Structure of the Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, MAGNESIUM ION, PHYCOERYTHROBILIN, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TZ9
 
 | |
7TZA
 
 | | Structure of PQS Response Protein PqsE in complex with N-(4-(3-neopentylureido)phenyl)-1H-indazole-7-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N-{4-[(2,2-dimethylpropyl)carbamamido]phenyl}-1H-indazole-7-carboxamide, ... | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PqsE Active Site as a Target for Small Molecule Antimicrobial Agents against Pseudomonas aeruginosa.
Biochemistry, 61, 2022
|
|
7U6G
 
 | |
6D0I
 
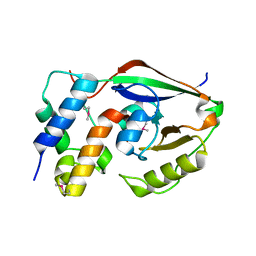 | | ParT: Prs ADP-ribosylating toxin bound to cognate antitoxin ParS. L48M ParT, SeMet-substituted complex. | | Descriptor: | GLYCEROL, ParS: COG5642 (DUF2384) antitoxin fragment, ParT: COG5654 (RES domain) toxin | | Authors: | Piscotta, F.J, Jeffrey, P.D, Link, A.J. | | Deposit date: | 2018-04-10 | | Release date: | 2019-01-09 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ParST is a widespread toxin-antitoxin module that targets nucleotide metabolism.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3BOH
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 1 with acetate (CDCA1-R1) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
4U6U
 
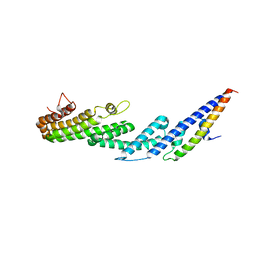 | |
2F1S
 
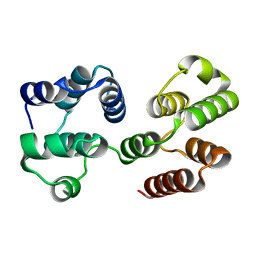 | | Crystal Structure of a Viral FLIP MC159 | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Li, F.-Y, Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2005-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Viral FLIP: INSIGHTS INTO FLIP-MEDIATED INHIBITION OF DEATH RECEPTOR SIGNALING.
J.Biol.Chem., 281, 2006
|
|
2F1Z
 
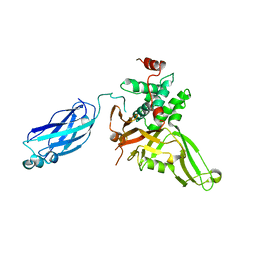 | | Crystal structure of HAUSP | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
2F1Y
 
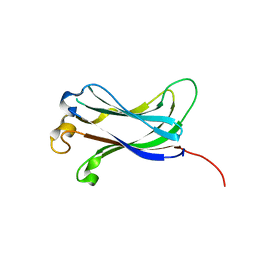 | |
2F1X
 
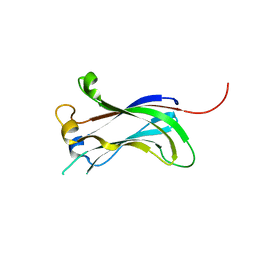 | |
7LTB
 
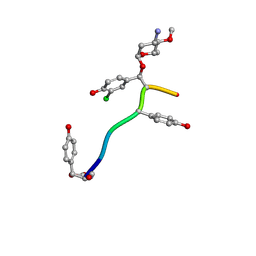 | | Crystal Structure of Keratinicyclin B | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, 3-ammonio-2,3,6-trideoxy-alpha-L-arabino-hexopyranose-(1-2)-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural and Functional Analysis of Keratinicyclin Reveals Synergistic Antibiosis with Vancomycin against Clostridium difficile
to be published
|
|
7LKC
 
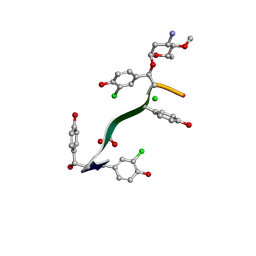 | | Crystal Structure of Keratinimicin A | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-02 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural and Functional Analysis of Keratinicyclin Reveals Synergistic Antibiosis with Vancomycin against Clostridium difficile
to be published
|
|
7N29
 
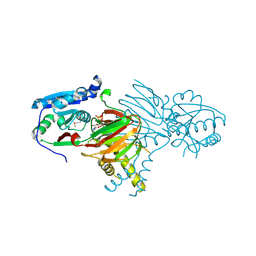 | | Structure of NAD kinase | | Descriptor: | NAD kinase 2, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Du, J, Estrella, M.A, Jeffrey, P.D, Korennykh, A.V. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human NADK2 reveals atypical assembly and regulation of NAD kinases from animal mitochondria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5BV0
 
 | |
5BUZ
 
 | |
5BV1
 
 | |
