4RMI
 
 | | Human Sirt2 in complex with SirReal1 and Ac-Lys-OTC peptide | | Descriptor: | Ac-Lys-OTC peptide, N-(5-benzyl-1,3-thiazol-2-yl)-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMJ
 
 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMH
 
 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
8OWZ
 
 | | Crystal structure of human Sirt2 in complex with a triazole-based SirReal | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[5-[[3-[[1-(2-methoxyethyl)-1,2,3-triazol-4-yl]methoxy]phenyl]methyl]-1,3-thiazol-2-yl]ethanamide, ... | | Authors: | Friedrich, F, Zhang, L, Schiedel, M, Einsle, O, Jung, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of First-in-Class Dual Sirt2/HDAC6 Inhibitors as Molecular Tools for Dual Inhibition of Tubulin Deacetylation.
J.Med.Chem., 66, 2023
|
|
5ANQ
 
 | | inhibitors of JumonjiC domain-containing histone demethylases | | Descriptor: | 2-{2-[(pyridin-3-ylmethyl)amino]pyrimidin-4-yl}pyridine-4-carboxylic acid, CHLORIDE ION, FE (II) ION, ... | | Authors: | Roatsch, M, Robaa, D, Pippel, M, Nettleship, J.E, Reddivari, Y, Bird, L.E, Hoffmann, I, Franz, H, Owens, R.J, Schuele, R, Flaig, R, Sippl, W, Jung, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substituted 2-(2-Aminopyrimidin-4-Yl)Pyridine-4-Carboxylates as Potent Inhibitors of Jumonjic Domain-Containing Histone Demethylases.
Fut.Med.Chem., 8, 2016
|
|
5DY4
 
 | | Crystal structure of human Sirt2 in complex with a brominated 2nd generation SirReal inhibitor and NAD+ | | Descriptor: | N-{5-[(7-bromonaphthalen-1-yl)methyl]-1,3-thiazol-2-yl}-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Aminothiazoles as Potent and Selective Sirt2 Inhibitors: A Structure-Activity Relationship Study.
J.Med.Chem., 59, 2016
|
|
5DY5
 
 | | Crystal structure of human Sirt2 in complex with a SirReal probe fragment | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Development of an Affinity Probe for Sirtuin 2.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4RMG
 
 | | Human Sirt2 in complex with SirReal2 and NAD+ | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
5D7Q
 
 | | Crystal structure of human Sirt2 in complex with ADPR and CHIC35 | | Descriptor: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5D7P
 
 | | Crystal structure of human Sirt2 in complex with ADPR and EX-243 | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5D7O
 
 | | Crystal structure of Sirt2-ADPR at an improved resolution | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, TRIETHYLENE GLYCOL, ZINC ION, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5D7N
 
 | | Crystal structure of human Sirt3 at an improved resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5YO9
 
 | |
5YOA
 
 | |
5YZU
 
 | |
5FUE
 
 | |
7R5B
 
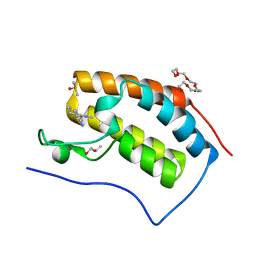 | | Crystal structure of BRD4(1) in complex with the inhibitor MPM2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1-(3-aminophenyl)-3-methyl-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrol-4-one, Bromodomain-containing protein 4, ... | | Authors: | Huegle, M. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A novel pan-selective bromodomain inhibitor for epigenetic drug design
Eur.J.Med.Chem., 249, 2023
|
|
7U6A
 
 | |
7U6B
 
 | |
7U69
 
 | |
7U3M
 
 | |
5NRW
 
 | |
5NU3
 
 | | Crystal structure of the human bromodomain of CREBBP bound to the inhibitor XDM-CBP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CREB-binding protein, SULFATE ION, ... | | Authors: | Huegle, M, Wohlwend, D. | | Deposit date: | 2017-04-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NU5
 
 | |
6XFP
 
 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | Descriptor: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
