6K7Q
 
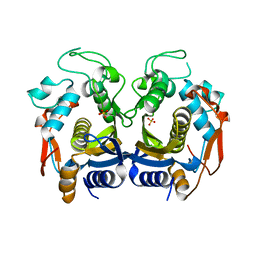 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
7N9H
 
 | |
3UR4
 
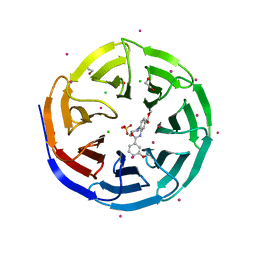 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
7EVR
 
 | | Crystal structure of hnRNP L RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L, SHI domain from Histone-lysine N-methyltransferase SETD2 | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7EVS
 
 | | Crystal structure of hnRNP LL RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L-like, SHI domain from Histone-lysine N-methyltransferase SETD2, SULFATE ION | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
4FMU
 
 | | Crystal structure of Methyltransferase domain of human SET domain-containing protein 2 Compound: Pr-SNF | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-(propylamino)hexanoic acid, Histone-lysine N-methyltransferase SETD2, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zeng, H, Ibanez, G, Zheng, W, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
3NY1
 
 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NY3
 
 | | Structure of the ubr-box of UBR2 in complex with N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR2, N-degron, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7ENQ
 
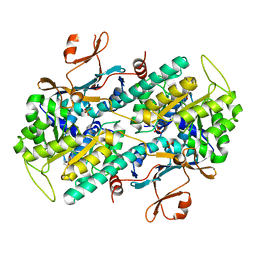 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
2FFL
 
 | |
4MI0
 
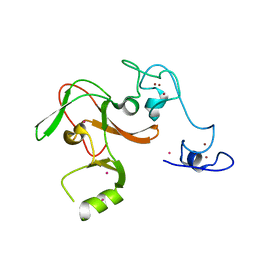 | | Human Enhancer of Zeste (Drosophila) Homolog 2(EZH2) | | Descriptor: | Histone-lysine N-methyltransferase EZH2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations.
Plos One, 8, 2013
|
|
7CKK
 
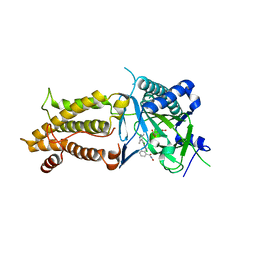 | | Structural complex of FTO bound with Dac51 | | Descriptor: | 2-{[2,6-dichloro-4-(3,5-dimethyl-1H-pyrazol-4-yl)phenyl]amino}-N-hydroxybenzamide, Alpha-ketoglutarate-dependent dioxygenase FTO, N-OXALYLGLYCINE | | Authors: | Yang, C, Gan, J. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance.
Cell Metab., 33, 2021
|
|
7DMP
 
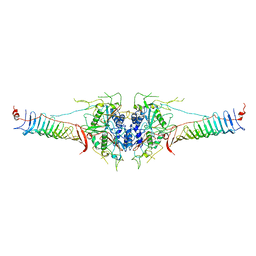 | | Mouse radial spoke complex | | Descriptor: | Radial spoke head 1 homolog, Radial spoke head protein 4 homolog A, Radial spoke head protein 9 homolog | | Authors: | Zheng, W, Cong, Y. | | Deposit date: | 2020-12-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct architecture and composition of mouse axonemal radial spoke head revealed by cryo-EM
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6LJA
 
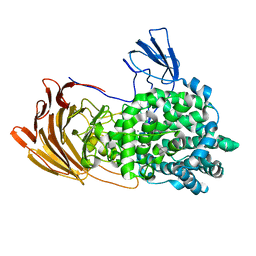 | | Crystal Structure of exoHep from Bacteroides intestinalis DSM 17393 complexed with disaccharide product | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
4LRV
 
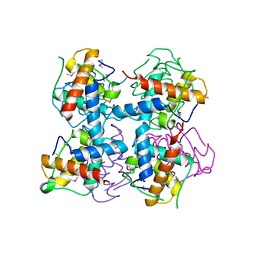 | | Crystal structure of DndE from Escherichia coli B7A involved in DNA phosphorothioation modification | | Descriptor: | DNA sulfur modification protein DndE | | Authors: | Hu, W, Wang, C.K, Liang, J.D, Zhang, T.L, Yang, M, Hu, Z.P, Wang, Z.J, Lan, W.X, Wu, H.M, Ding, J.P, Wu, G, Deng, Z.X, Cao, C. | | Deposit date: | 2013-07-21 | | Release date: | 2013-08-28 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification
Cell Res., 22, 2012
|
|
2GFS
 
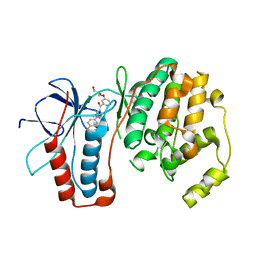 | | P38 Kinase Crystal Structure in complex with RO3201195 | | Descriptor: | Mitogen-Activated Protein Kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Harris, S.F, Bertrand, J, Villasenor, A. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Discovery of S-[5-Amino-1-(4-fluorophenyl)-1H-pyrazol-4-yl]-[3-(2,3-dihydroxypropoxy)phenyl]-methanone (RO3201195), and Orally Bioavailable and Highly Selective Inhibitor of p38 Map Kinase
J.Med.Chem., 49, 2006
|
|
6LJL
 
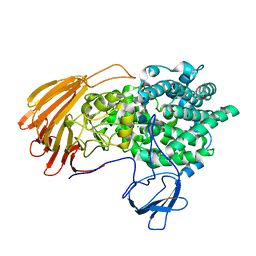 | | Crystal Structure of exoHep-Y390A/H555A complexed with a tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
6KI6
 
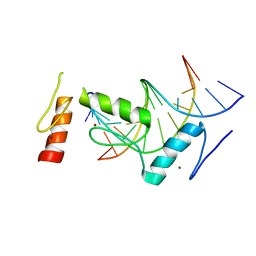 | | Crystal structure of BCL11A in complex with gamma-globin -115 HPFH region | | Descriptor: | B-cell lymphoma/leukemia 11A, DNA (5'-D(*AP*TP*AP*TP*TP*GP*GP*TP*CP*AP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*GP*AP*CP*CP*AP*AP*TP*A)-3'), ... | | Authors: | Li, F.D, Yang, Y, Shi, Y.Y. | | Deposit date: | 2019-07-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the recognition of gamma-globin gene promoter by BCL11A.
Cell Res., 29, 2019
|
|
7DTK
 
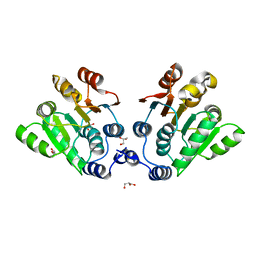 | |
7DTJ
 
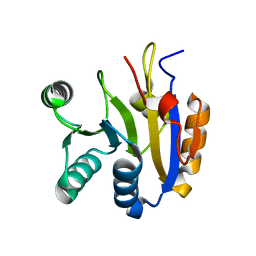 | |
8HL8
 
 | | Crystal structrue of MtdL R257K mutant | | Descriptor: | MANGANESE (II) ION, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-11-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
7C1P
 
 | | Crystal structure of the starter condensation domain of the rhizomide synthetase RzmA mutant H140V, R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1S
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA and Leu-SNAC | | Descriptor: | Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A, S-(2-acetamidoethyl) (2S)-2-azanyl-4-methyl-pentanethioate | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7W6K
 
 | | Cryo-EM structure of GmALMT12/QUAC1 anion channel | | Descriptor: | GmALMT12/QUAC1 | | Authors: | Qin, L, Tang, L.H, Xu, J.S, Zhang, X.H, Zhu, Y, Sun, F, Su, M, Zhai, Y.J, Chen, Y.H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure and electrophysiological characterization of ALMT from Glycine max reveal a previously uncharacterized class of anion channels.
Sci Adv, 8, 2022
|
|
