4UR9
 
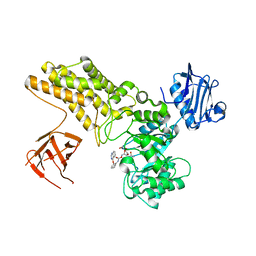 | | Structure of ligand bound glycosylhydrolase | | Descriptor: | 4-ethoxyquinazoline, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, ... | | Authors: | Darby, J.F, Landstroem, J, Roth, C, He, Y, Schultz, M, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2014-06-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Selective Small-Molecule Activators of a Bacterial Glycoside Hydrolase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5FL0
 
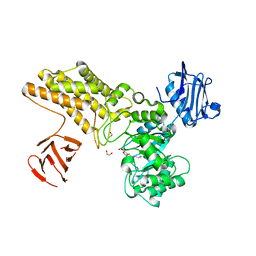 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-(butylamino)-5-(hydroxymethyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d] [1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
7OU8
 
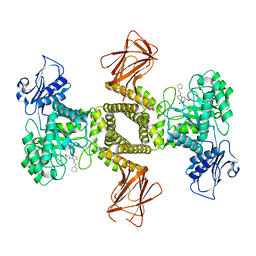 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, O-GlcNAcase BT_4395, ... | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
5MI4
 
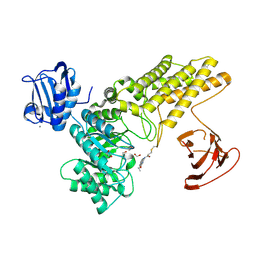 | | BtGH84 mutant with covalent modification by MA3 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, O-GlcNAcase BT_4395, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
5OXD
 
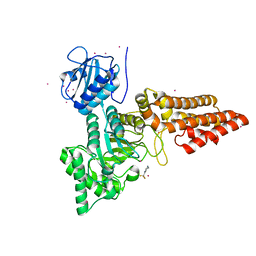 | |
2WZI
 
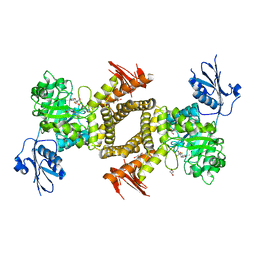 | | BtGH84 D243N in complex with 5F-oxazoline | | Descriptor: | (3AS,5S,6S,7R,7AR)-5-FLUORO-5-(HYDROXYMETHYL)-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]OXAZOLE-6,7-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualizing the Reaction Coordinate of an O-Glcnac Hydrolase
J.Am.Chem.Soc., 132, 2010
|
|
2X0Y
 
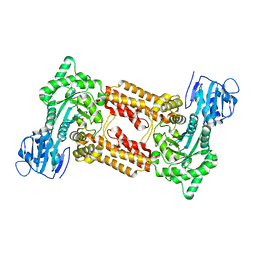 | | Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds | | Descriptor: | 7-[(2S)-2,3-DIHYDROXYPROPYL]-1,3-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE, O-GLCNACASE NAGJ | | Authors: | Dorfmueller, H.C, van Aalten, D.M.F. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening-Based Discovery of Drug-Like O-Glcnacase Inhibitor Scaffolds
FEBS Lett., 584, 2010
|
|
5FKY
 
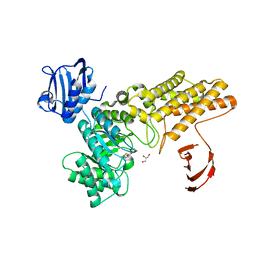 | | Structure of a hydrolase bound with an inhibitor | | Descriptor: | (3aR,5R,6S,7R,7aR)-2-amino-5-(hydroxymethyl)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, GLYCEROL, O-GLCNACASE BT_4395 | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
2XM2
 
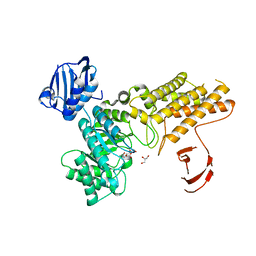 | | BtGH84 in complex with LOGNAc | | Descriptor: | GLYCEROL, N-acetylglucosaminono-1,5-lactone (Z)-oxime, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibition of a Bacterial O-Glcnacase Homologue by Lactone and Lactam Derivatives: Structural, Kinetic and Thermodynamic Analyses.
Amino Acids, 40, 2011
|
|
5MI7
 
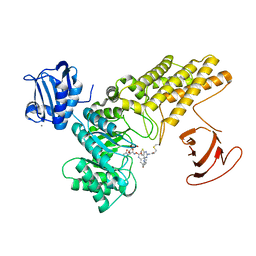 | | BtGH84 mutant with covalent modification by MA4 in complex with PUGNAc | | Descriptor: | CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, O-GlcNAcase BT_4395, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
5MI5
 
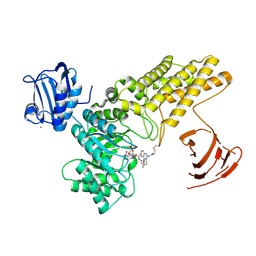 | | BtGH84 mutant with covalent modification by MA3 in complex with PUGNAc | | Descriptor: | CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, O-GlcNAcase BT_4395, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
5MI6
 
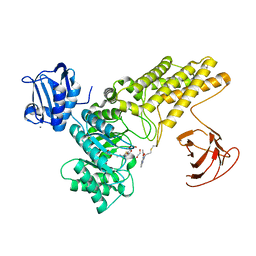 | | BtGH84 mutant with covalent modification by MA3 in complex with Thiamet G | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
7K41
 
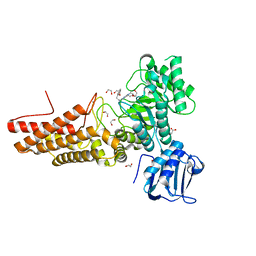 | | Bacterial O-GlcNAcase (OGA) with compound | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methylpiperidin-1-yl)-N-(2-phenylethyl)pyrimidin-2-amine, ACETATE ION, ... | | Authors: | Lane, W, Tjhen, R, Snell, G, Sang, B. | | Deposit date: | 2020-09-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Novel and Brain-Penetrant O -GlcNAcase Inhibitor via Virtual Screening, Structure-Based Analysis, and Rational Lead Optimization.
J.Med.Chem., 64, 2021
|
|
7KHV
 
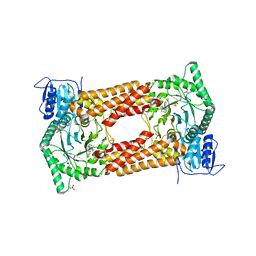 | | CpOGA IN COMPLEX WITH LIGAND 54 | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-(5-{[6-(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)-2,6-diazaspiro[3.4]octan-2-yl]methyl}-1,3-thiazol-2-yl)acetamide, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Diazaspirononane Nonsaccharide Inhibitors of O-GlcNAcase (OGA) for the Treatment of Neurodegenerative Disorders.
J.Med.Chem., 63, 2020
|
|
2CBJ
 
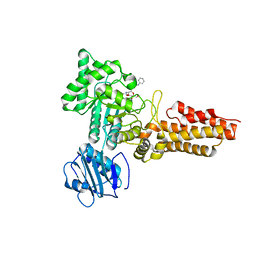 | | Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase in complex with PUGNAc | | Descriptor: | CHLORIDE ION, HYALURONIDASE, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Rao, F.V, Dorfmueller, H.C, Villa, F, Allwood, M, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis.
EMBO J., 25, 2006
|
|
2J62
 
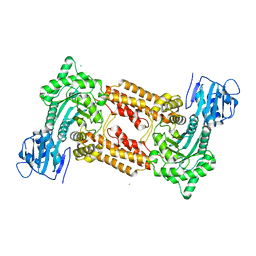 | | Structure of a bacterial O-glcnacase in complex with glcnacstatin | | Descriptor: | CHLORIDE ION, N-[(5R,6R,7R,8S)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDIN-8-YL]-2-METHYLPROPANAMIDE, O-GlcNAcase NagJ | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, Shepherd, S.M, Shpiro, N.A, van Aalten, D.M.F. | | Deposit date: | 2006-09-22 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels.
J. Am. Chem. Soc., 128, 2006
|
|
2CBI
 
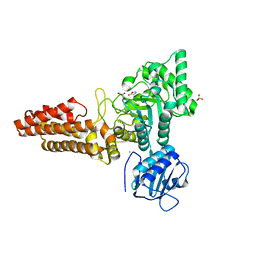 | | Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase | | Descriptor: | CHLORIDE ION, GAMMA-BUTYROLACTONE, GLYCEROL, ... | | Authors: | Rao, F.V, Dorfmueller, H.C, Villa, F, Allwood, M, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2006-01-04 | | Release date: | 2006-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis.
EMBO J., 25, 2006
|
|
2CHN
 
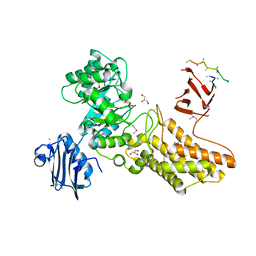 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity- NAG-thiazoline complex | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
2J47
 
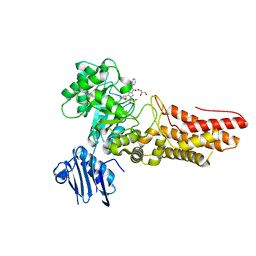 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with a imidazole-pugnac hybrid inhibitor | | Descriptor: | (5R,6R,7R,8S)-8-(ACETYLAMINO)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-N-PHENYL-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDINE-2-CARBOXAMIDE, GLUCOSAMINIDASE, GLYCEROL | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2006-08-25 | | Release date: | 2006-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Inhibition of O-Glcnacase by a Gluco-Configured Nagstatin and a Pugnac-Imidazole Hybrid Inhibitor
Chem. Commun., 42, 2006
|
|
2J4G
 
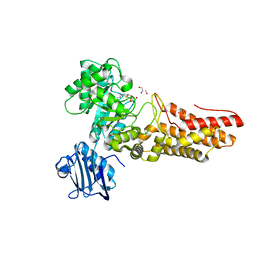 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor | | Descriptor: | (3AR,5R,6S,7R,7AR)-5-(HYDROXYMETHYL)-2-PROPYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of Pugnac and Nag-Thiazoline as Transition State Analogues for Human O-Glcnacase: Mechanistic and Structural Insights Into Inhibitor Selectivity and Transition State Poise.
J.Am.Chem.Soc., 129, 2007
|
|
5FL1
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
2CHO
 
 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity | | Descriptor: | ACETATE ION, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
2JIW
 
 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine | | Descriptor: | N-[(1S,2R,5R,6R)-2-AMINO-5,6-DIHYDROXY-4-(HYDROXYMETHYL)CYCLOHEX-3-EN-1-YL]ACETAMIDE, O-GLCNACASE BT_4395 | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A 1-Acetamido Derivative of 6-Epi-Valienamine: An Inhibitor of a Diverse Group of Beta-N-Acetylglucosaminidases.
Org.Biomol.Chem., 5, 2007
|
|
6PV5
 
 | | Structure of CpGH84B | | Descriptor: | 1,2-ETHANEDIOL, Putative O-GlcNAcase nagJ | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
6PWI
 
 | | Structure of CpGH84D | | Descriptor: | 1,2-ETHANEDIOL, Putative hyaluronoglucosaminidase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
