7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
8BWY
 
 | | In situ outer dynein arm from Chlamydomonas reinhardtii in a pre-power stroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Calmodulin, ... | | Authors: | Zimmermann, N.E.L, Noga, A, Obbineni, J.M, Ishikawa, T. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (38 Å) | | Cite: | ATP-induced conformational change of axonemal outer dynein arms revealed by cryo-electron tomography.
Embo J., 42, 2023
|
|
7KEK
 
 | | Structure of the free outer-arm dynein in pre-parallel state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein alpha heavy chain, ... | | Authors: | Rao, Q, Zhang, K. | | Deposit date: | 2020-10-11 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structures of outer-arm dynein array on microtubule doublet reveal a motor coordination mechanism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7WGE
 
 | | Human NLRP1 complexed with thioredoxin | | Descriptor: | MAGNESIUM ION, NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2021-12-28 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for thioredoxin-mediated suppression of NLRP1 inflammasome.
Nature, 622, 2023
|
|
7QNG
 
 | | Structure of a MHC I-Tapasin-ERp57 complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Mueller, I.K, Thomas, C, Trowitzsch, S, Tampe, R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity.
Nat Commun, 13, 2022
|
|
7QPD
 
 | | Structure of the human MHC I peptide-loading complex editing module | | Descriptor: | Beta-2-microglobulin, Calreticulin, HLA class I histocompatibility antigen, ... | | Authors: | Domnick, A, Susac, L, Trowitzsch, S, Thomas, C, Tampe, R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Molecular basis of MHC I quality control in the peptide loading complex.
Nat Commun, 13, 2022
|
|
7MOQ
 
 | | The structure of the Tetrahymena thermophila outer dynein arm on doublet microtubule | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Docking complex 1 protein, ... | | Authors: | Kubo, S, Yang, S.K, Ichikawa, M, Bui, K.H. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Remodeling and activation mechanisms of outer arm dyneins revealed by cryo-EM.
Embo Rep., 22, 2021
|
|
6Y4Z
 
 | | The crystal structure of human MACROD2 in space group P43212 | | Descriptor: | L(+)-TARTARIC ACID, Thioredoxin 1,ADP-ribose glycohydrolase MACROD2 | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6ENY
 
 | | Structure of the human PLC editing module | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Calreticulin, ... | | Authors: | Trowitzsch, S, Januliene, D, Blees, A, Moeller, A, Tampe, R. | | Deposit date: | 2017-10-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure of the human MHC-I peptide-loading complex.
Nature, 551, 2017
|
|
8PKO
 
 | | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Hitchman, C.J, Lia, A, Bayo, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer).
To Be Published
|
|
5AYK
 
 | | Crystal structure of ERdj5 form I | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CHLORIDE ION, DnaJ homolog subfamily C member 10 | | Authors: | Watanabe, S, Maegawa, K, Inaba, K. | | Deposit date: | 2015-08-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
5AYL
 
 | | Crystal structure of ERdj5 form II | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, DnaJ homolog subfamily C member 10 | | Authors: | Watanabe, S, Maegawa, K, Inaba, K. | | Deposit date: | 2015-08-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
7K58
 
 | |
6N7W
 
 | | Structure of bacteriophage T7 leading-strand DNA polymerase (D5A/E7A)/Trx in complex with a DNA fork and incoming dTTP (from multiple lead complexes) | | Descriptor: | DNA (25-MER), DNA (77-MER), DNA-directed DNA polymerase, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
1ZYQ
 
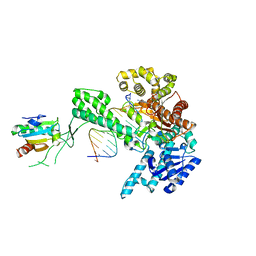 | | T7 DNA polymerase in complex with 8oG and incoming ddATP | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*CP*(8OG)P*CP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(DDG))-3', ... | | Authors: | Brieba, L.G, Kokoska, R.J, Bebenek, K, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2005-06-10 | | Release date: | 2005-11-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A lysine residue in the fingers subdomain of t7 DNA polymerase modulates the miscoding potential of 8-oxo-7,8-dihydroguanosine.
Structure, 13, 2005
|
|
8EKY
 
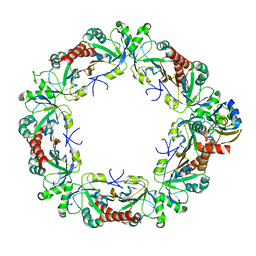 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EOJ
 
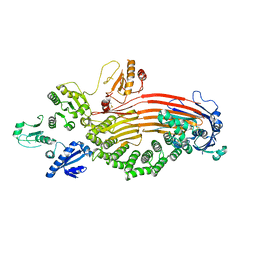 | | Microsomal triglyceride transfer protein | | Descriptor: | Microsomal triglyceride transfer protein large subunit, Protein disulfide-isomerase | | Authors: | Zhang, Z. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
6TMG
 
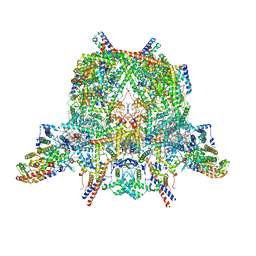 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, membrane region model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ATPTG1, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
7P9D
 
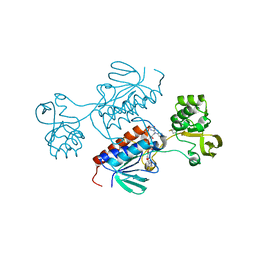 | | Crystal structure of Chlamydomonas reinhardtii NADPH Dependent Thioredoxin Reductase 1 domain | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Singh, R.K, Marchetti, G.M, Hippler, M, Kuemmel, D. | | Deposit date: | 2021-07-27 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis revealed a novel conformation of the NTRC reductase domain from Chlamydomonas reinhardtii.
J.Struct.Biol., 214, 2021
|
|
7P9E
 
 | |
6I7S
 
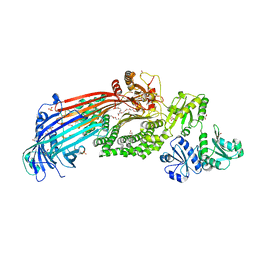 | | Microsomal triglyceride transfer protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Biterova, E, Isupov, M.N, Keegan, R.M, Lebedev, A.A, Ruddock, L.W. | | Deposit date: | 2018-11-17 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human microsomal triglyceride transfer protein.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7YSI
 
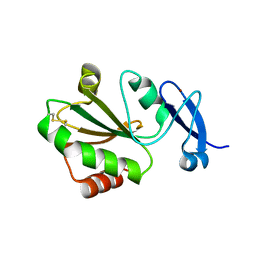 | | Crystal structure of thioredoxin 2 | | Descriptor: | Thiol disulfide reductase thioredoxin, ZINC ION | | Authors: | Chang, Y.J, Park, H.H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Comparison of the structure and activity of thioredoxin 2 and thioredoxin 1 from Acinetobacter baumannii.
Iucrj, 10, 2023
|
|
8AON
 
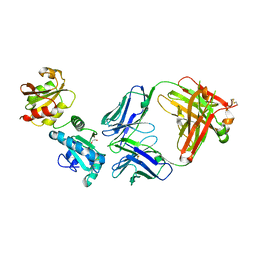 | |
7ZSC
 
 | | Crystal structure of the heterodimeric human C-P4H-II with truncated alpha subunit (C-P4H-II delta281) | | Descriptor: | Prolyl 4-hydroxylase subunit alpha-2, Protein disulfide-isomerase, SULFATE ION | | Authors: | Lebedev, A, Koski, M.K, Wierenga, R.K, Murthy, A.V, Sulu, R. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the collagen prolyl 4-hydroxylase (C-P4H) catalytic domain complexed with PDI: Toward a model of the C-P4H alpha 2 beta 2 tetramer.
J.Biol.Chem., 298, 2022
|
|
