Find the service you need
Choose a keyword listed below or input keywords into the textbox at the right of the keyword list. The brief explanation of the matched services will be displayed.
- Click the 'Show all services' button to display the explanation for all services.
- Input some keywords into the 'Word Search Box' to narrow down the search results.
 | PDB Deposition PDB Deposition is a new deposition tool developed by the wwPDB partners. It supports X-ray, NMR and EM structures. PDB, BMRB, EMDB, deposition, NMR, electron microscopy |
 | non-PDB format compatible non-PDB format compatible are entries which exceed the limitations of the PDB format for a few reasons and cannot be represented in a regular single PDB format file. PDB, 3D structure, search |
 | Group Depositions Group Depositions are sets of related PDB entries deposited at the same time. PDB entries belonging to each group are available by clicking on the Group ID. PDB, 3D structure, search |
 | Chemical Component entries All Chemical Component entries defined in Chemical Component dictionary are listed. chemical component, search, 3D structure, viewer |
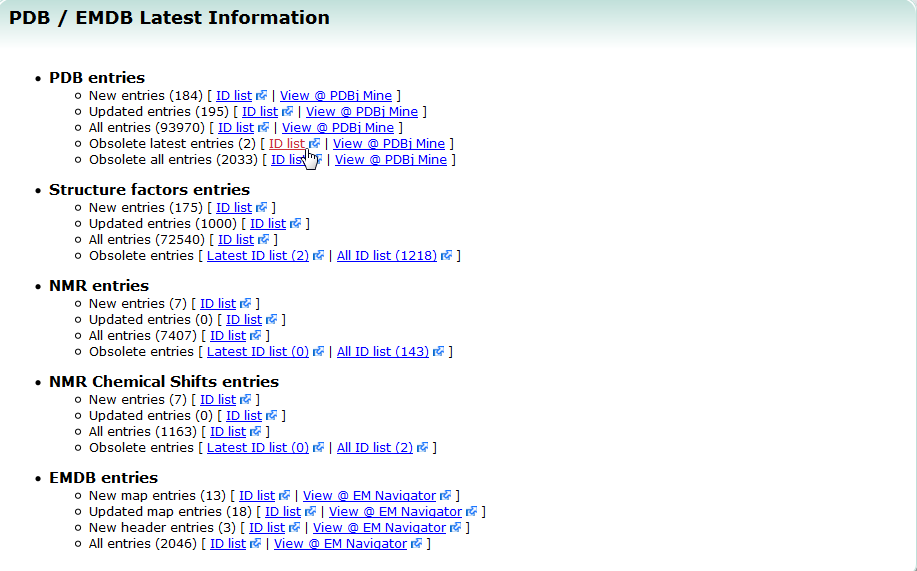 | Latest entries Latest entries provides the list of latest released entries including both newly registered entries and updated entries. Each entry ID is linked to the PDBj Mine page. All the entry ID list in PDB is also available. PDB, search |
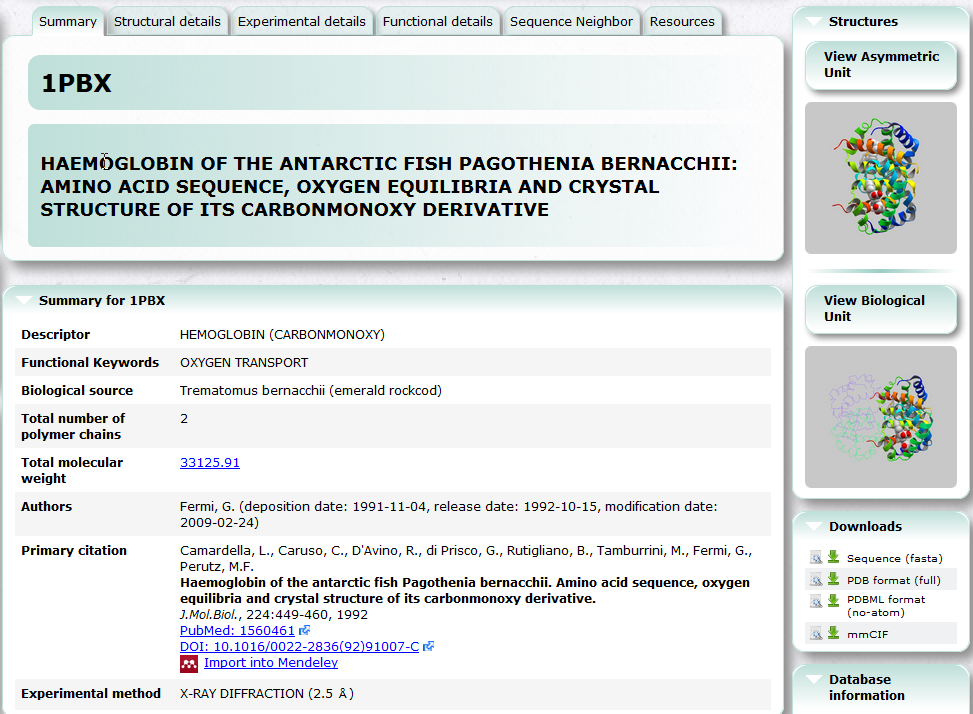 | PDBj Mine PDBj Mine is a new generic interface to search PDB entries by PDB ID and keyword etc. See also Basic Usage. PDB, 3D structure, search, chemical component |
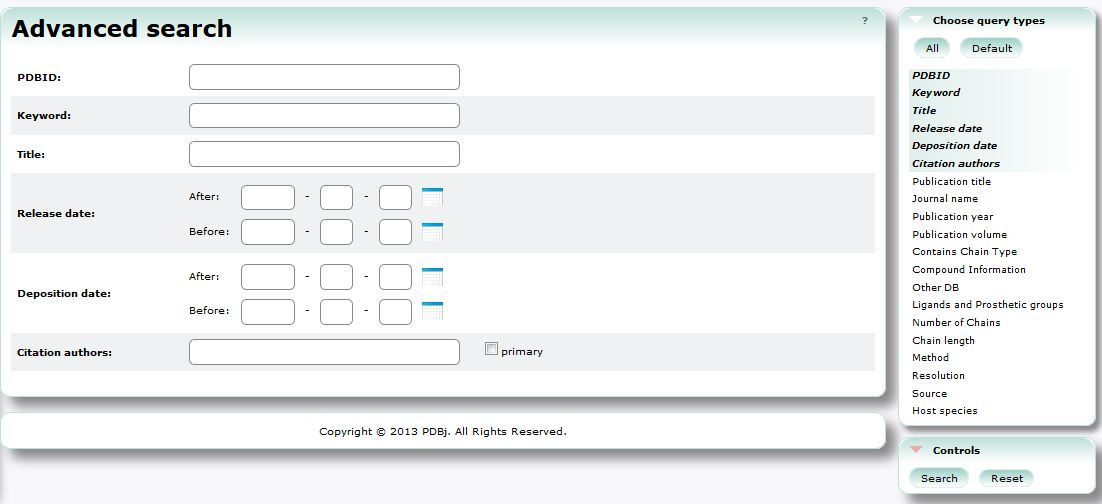 | Advanced Search (PDBj Mine) Advanced Search is an advanced interface to search PDB entries by a combination of various conditions. PDB, 3D structure, search, chemical component |
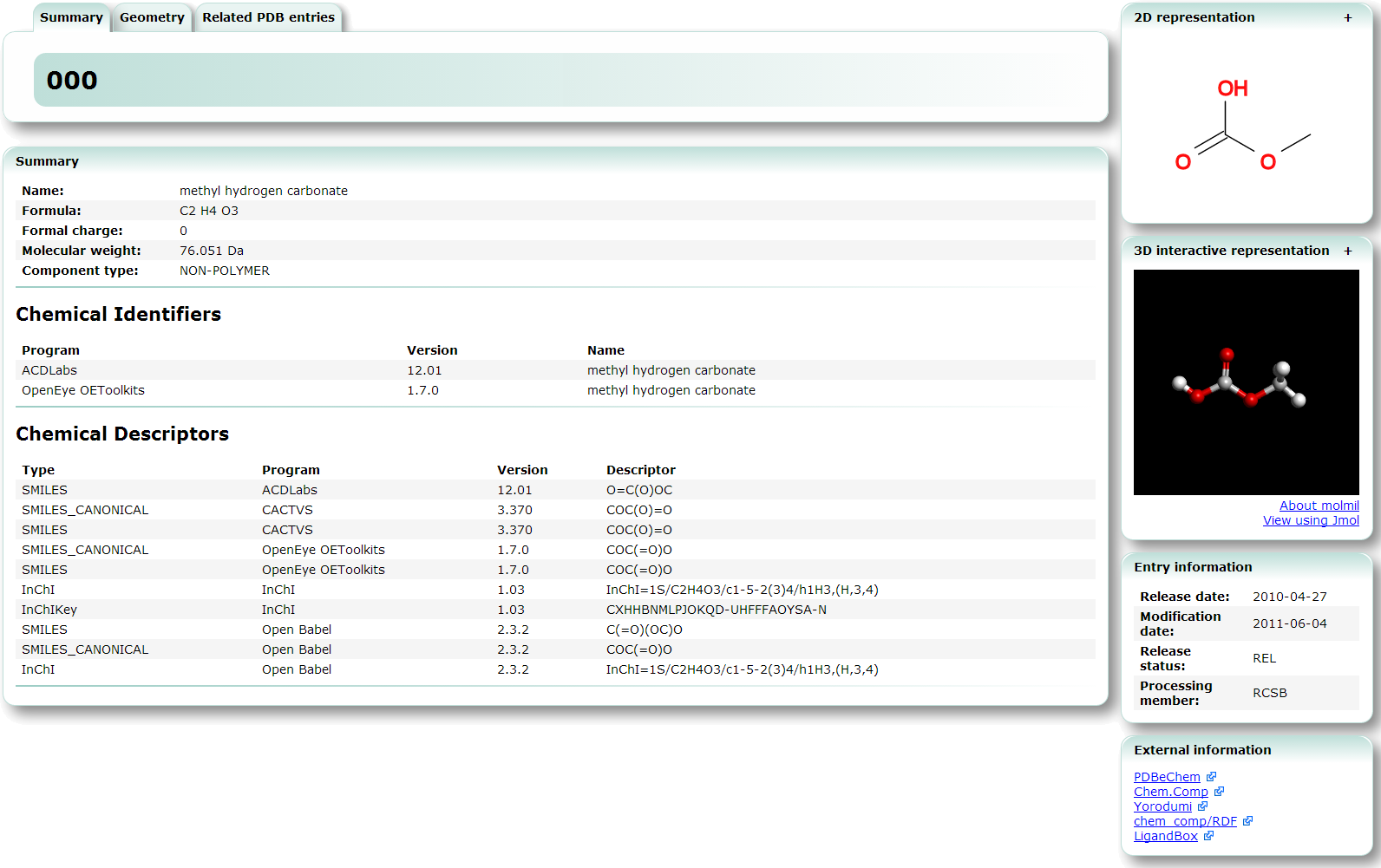 | Chemie search Chemie search can be used to search the chemical component database which is part of the PDB. chemical component, search, 3D structure, viewer |
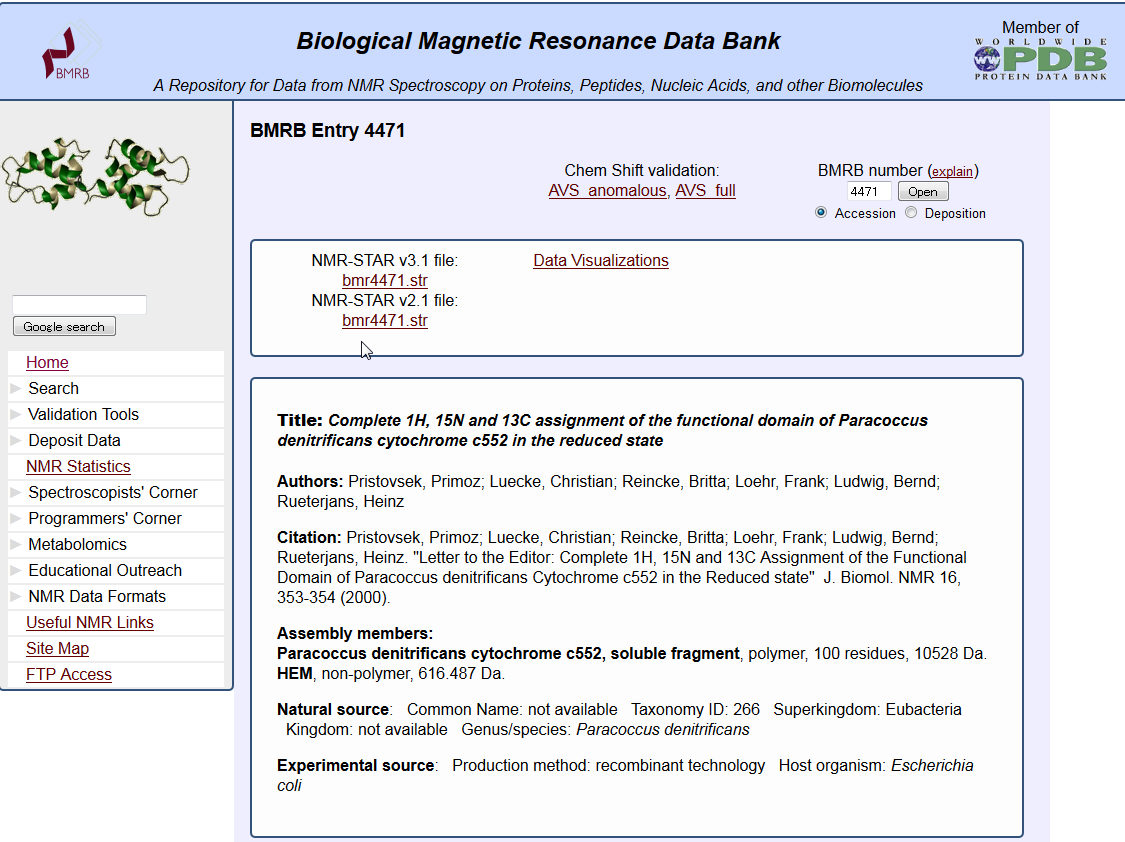 | NMR data search (BMRB) NMR data search (BMRB) is a service to search BMRB entries in the biomolecule NMR database BMRB. BMRB, NMR |
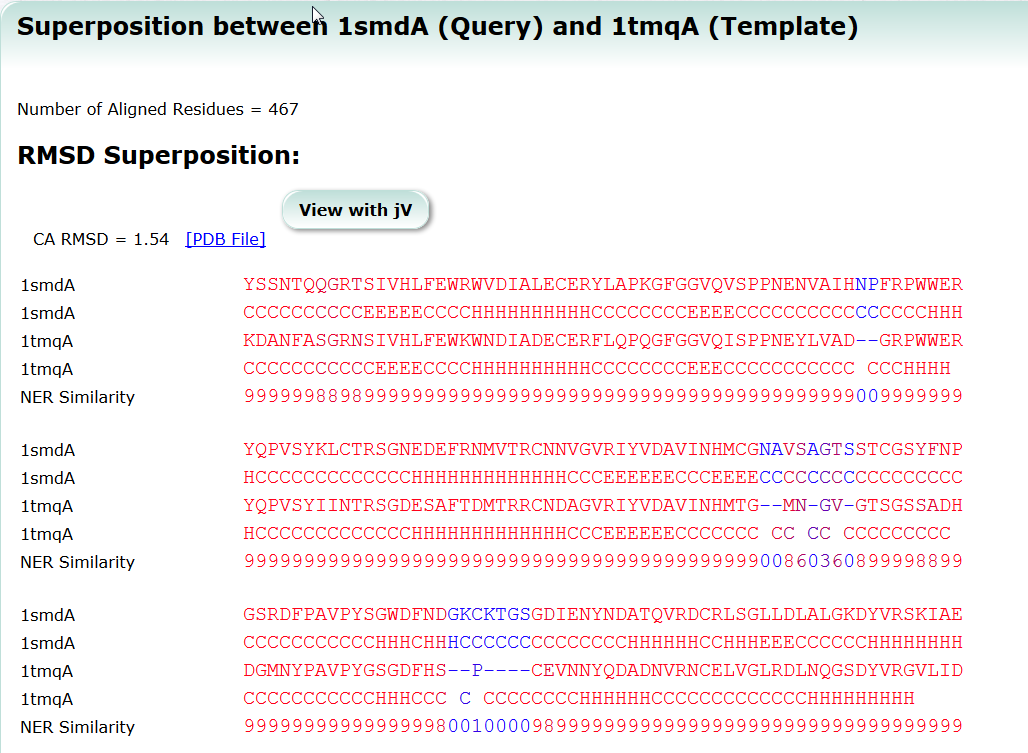 | Sequence Navigator Sequence Navigator is a tool to look for molecules with similar sequences. sequence, search, similarity |
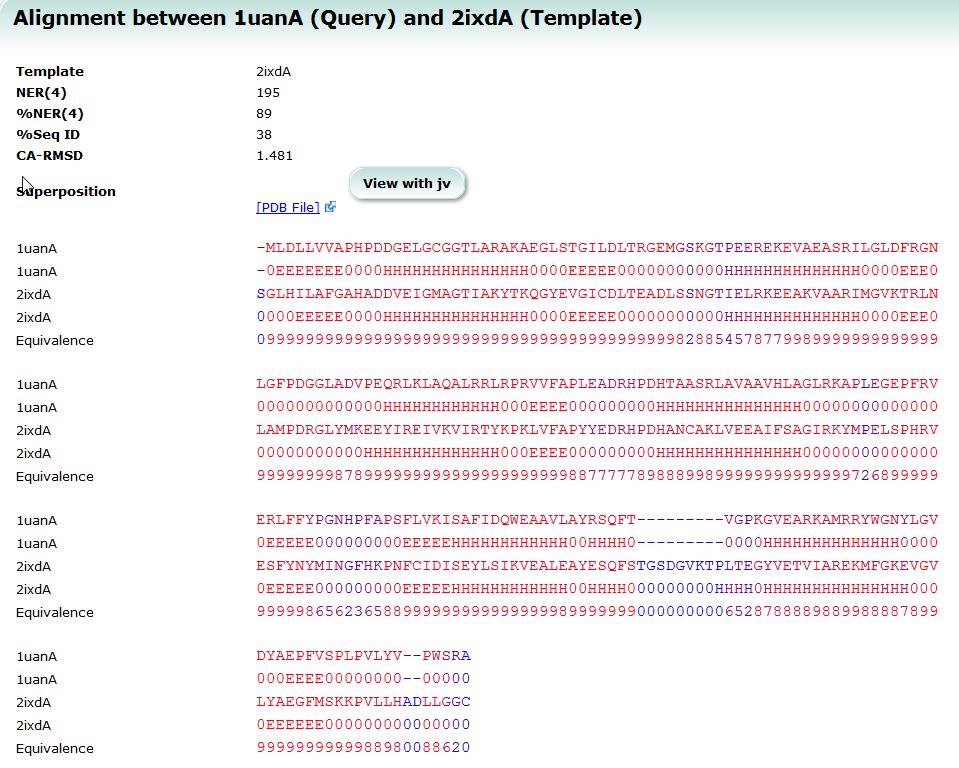 | Structure Navigator Structure Navigator is a tool to look for molecules with similar structures. 3D structure, search, similarity |
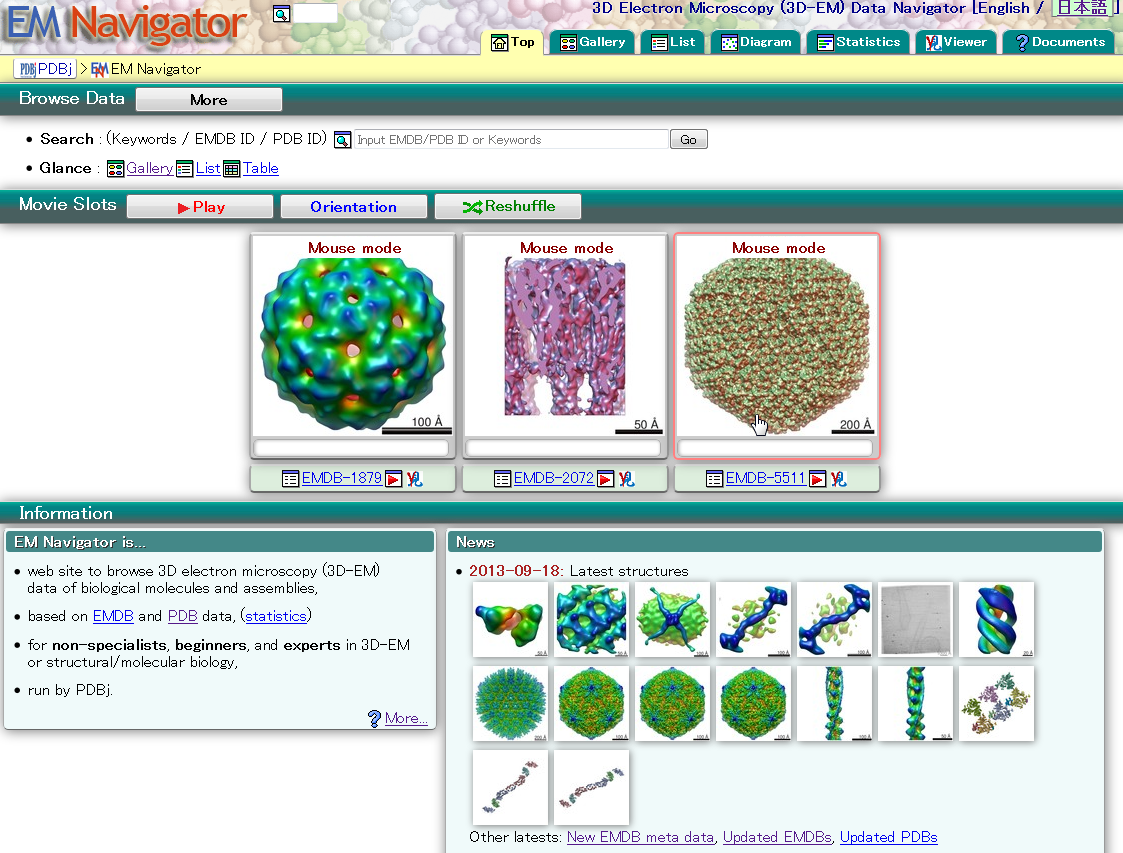 | EM Navigator EM Navigator is a data browser for biological 3D electron microscopy. EMDB, electron microscopy, search, 3D structure |
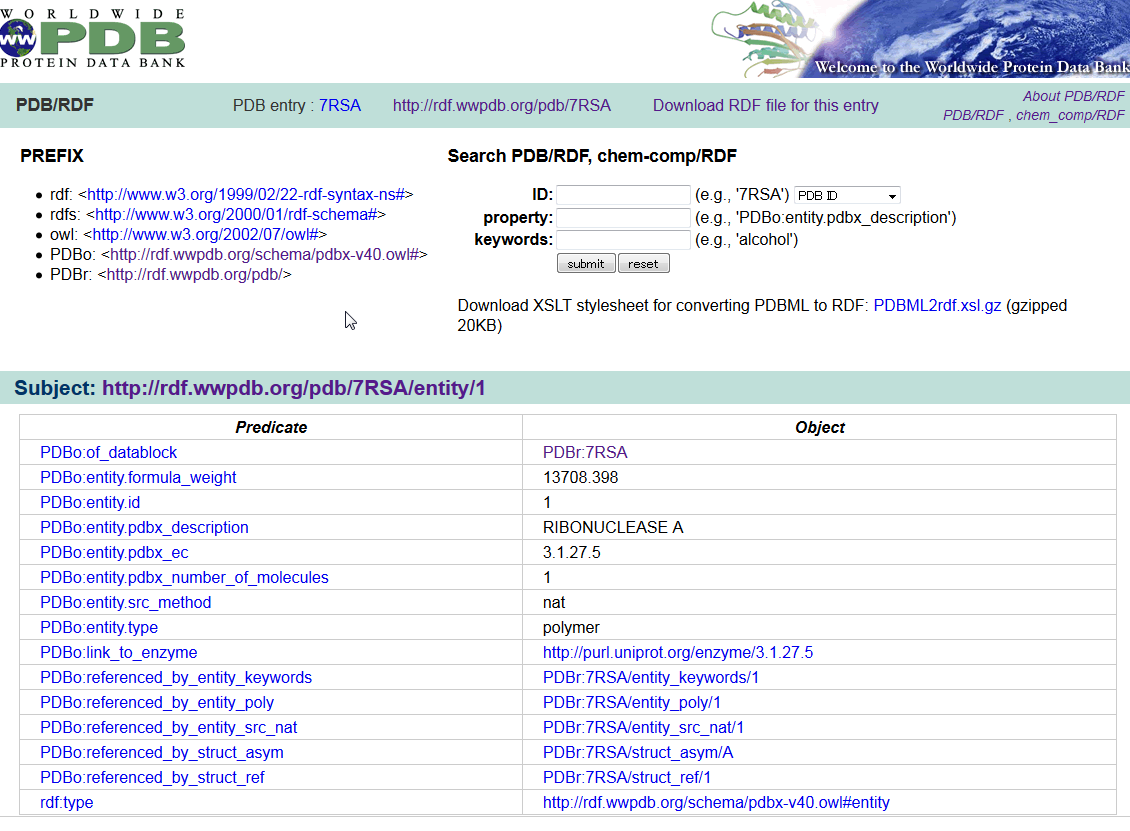 | wwPDB/RDF wwPDB/RDF is a collection of PDB data in the Resource Description Framework (RDF) format. The RDF format is the standard format for the Semantic Web. An ontology defined in the Web Ontology Language (OWL) is also provided for the PDB/RDF, which is a straightforward translation of the PDB mmCIF Exchange Dictionary. PDB, chemical component, RDF |
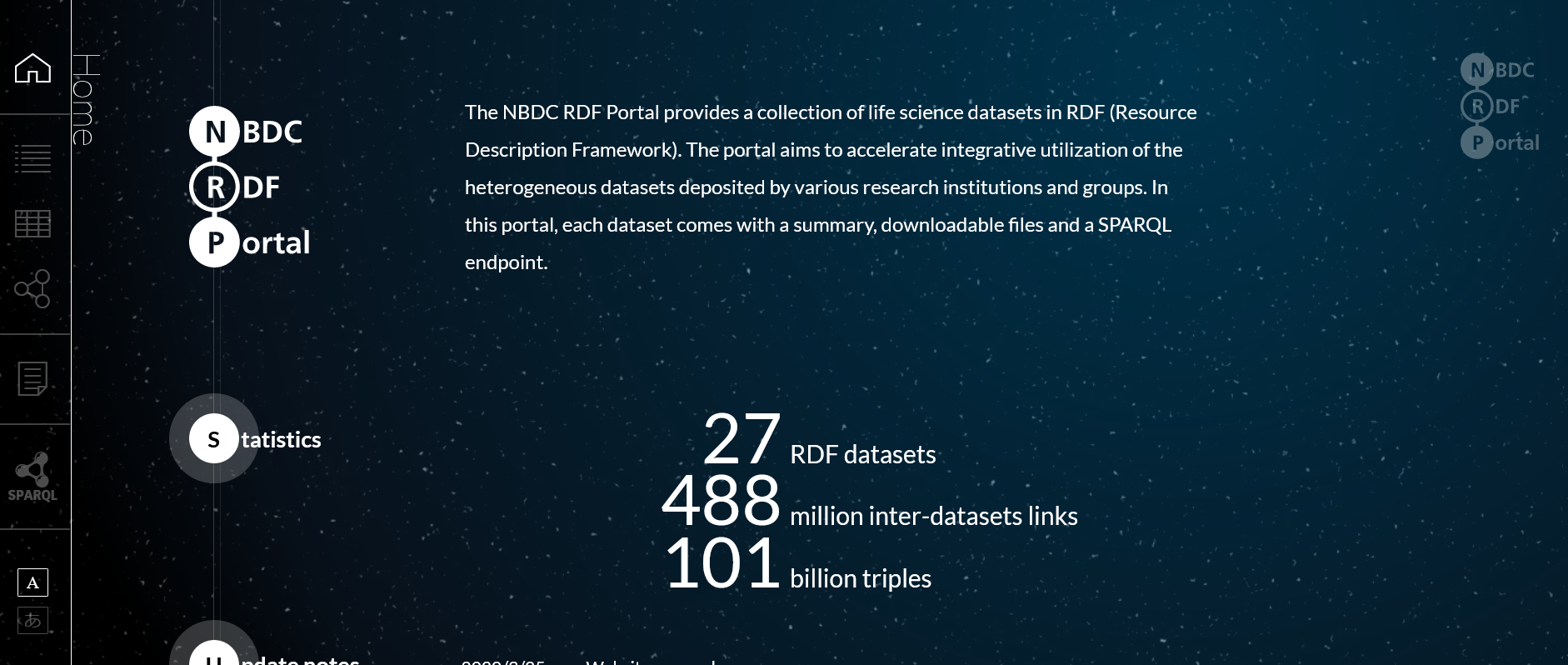 | RDF Portal RDF Portal is SPARQL endpoints for comprehensive RDF graphs, which contains RDF graphs for PDB core, wwPDB validation reports, and SIFTS datasets, respectively. RDF graph search using SPARQL is possible on the site. RDF, SPARQL, gene, genome, sequence, disease, drug |
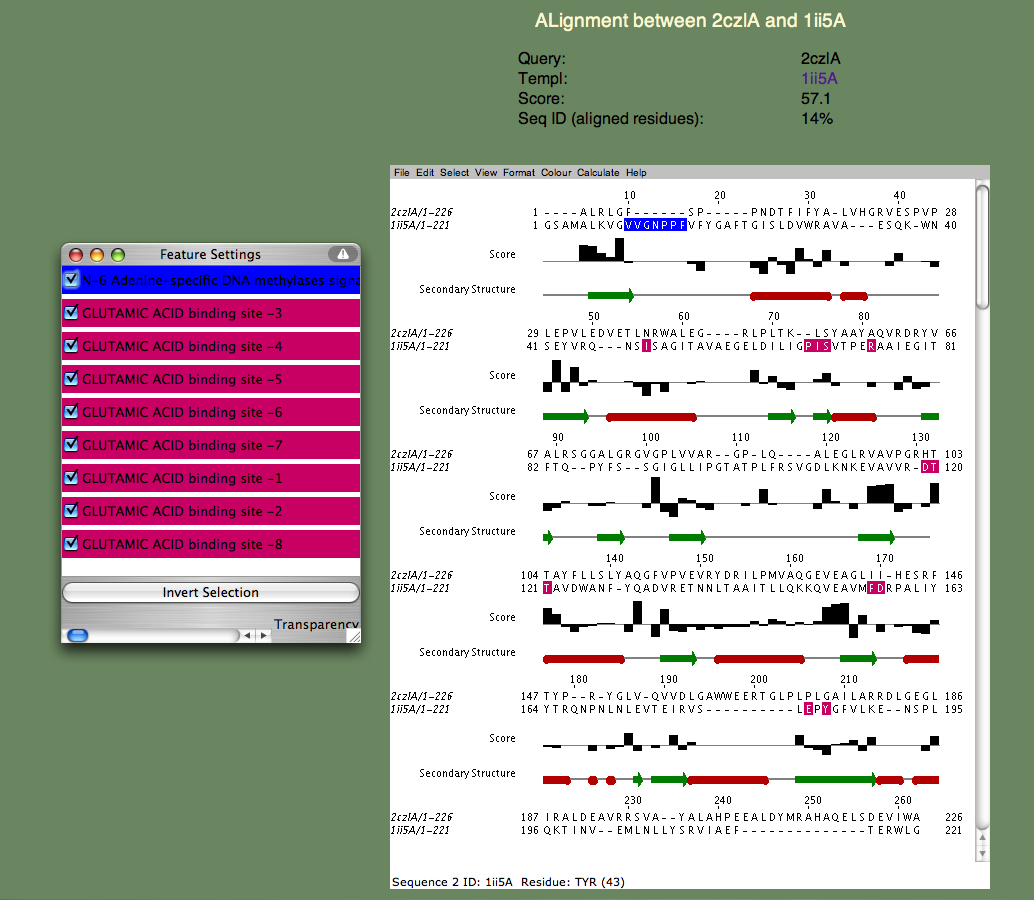 | SeSAW SeSAW (Sequence-Derived Structural Alignment Weights) provides functional annotations that identifies conserved sequence and structural motifs in query proteins. See also SeSAW Tutorial. sequence, 3D structure, function prediction |
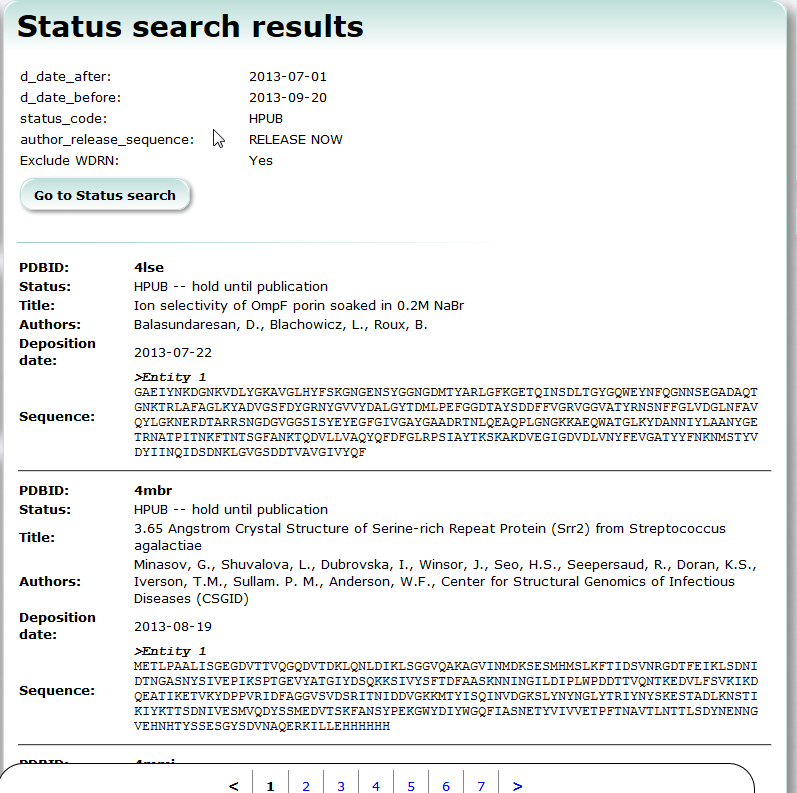 | Status Search Status Search is a service to search PDB entries which are already deposited but have not been released yet. PDB, search |
 | jV (PDBj Viewer) jV is an interactive 3D viewer program to visualize structure of protein and nucleic acid molecules. This program is also possible to use jV3 on a web browser as a Java applet. viewer, 3D structure, PDB |
 | Molmil Molmil is a new high performance molecular viewer developed by PDBj using modern web technology for optimal integration and user experience. It can be used for visualization of proteins, nucleic acids and chemical compounds present in PDB (flat), mmCIF, PDBML and the PDBj developed format mmJSON. It also supports saving PNG images of high resolution and runs inside modern web-browsers without any need for plugins. 3D structure, viewer |
 | Yorodumi Yorodumi is a website to easily watch, move, rotate, learn, and enjoy 3D structures of biological molecules. PDB, EMDB, chemical component, 3D structure, viewer |
 | ASH ASH is a program to perform three-dimensional structure alignment. From the ASH homepage you can run the program over the web or download the software for free. similarity, search, 3D structure |
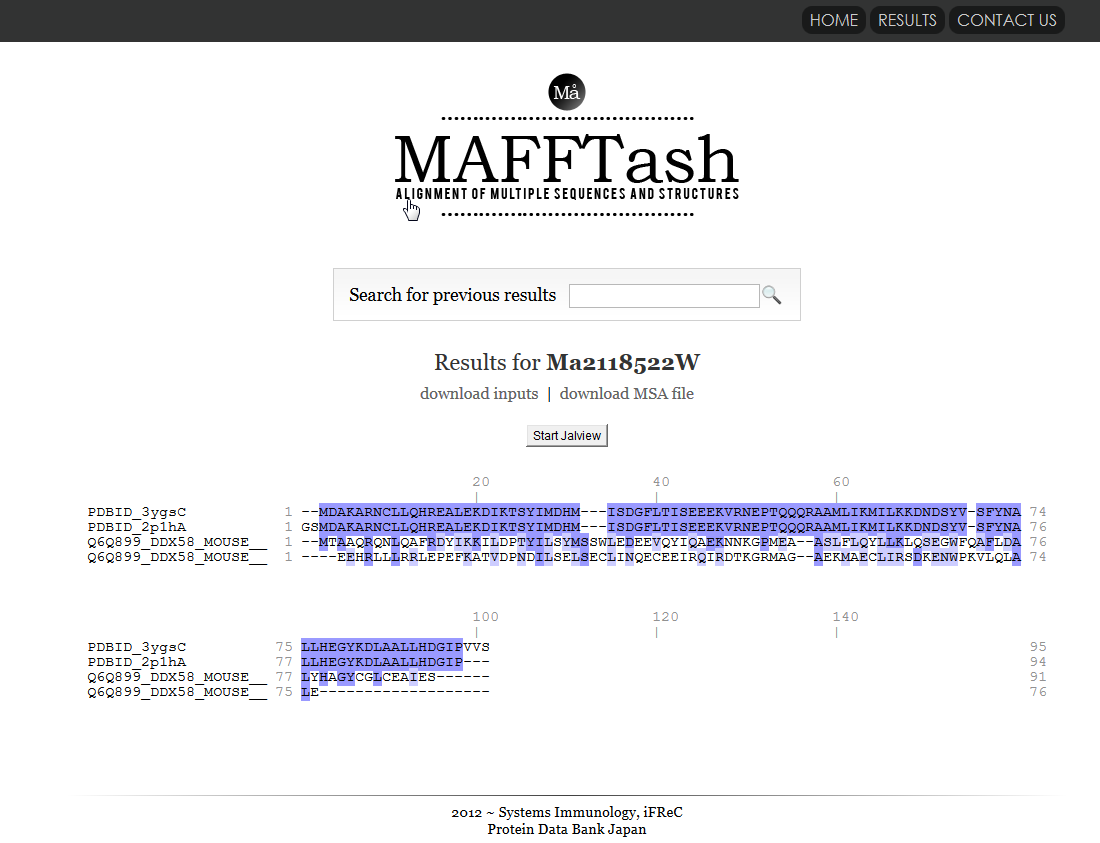 | MAFFTash MAFFTash is a tool that calculates multple sequence alignments from sequences and structures. similarity, search, 3D structure, sequence |
 | NMRToolBox NMRToolBox is composed of various tools for spectral assignment and analysis NMR, search |
 | CRNPRED CRNPRED is a web-based service that predicts one-dimensional protein structures including secondary structures, contact numbers, and residue-wise contact orders from amino acid sequence. secondary structure, 3D structure, structure prediction |
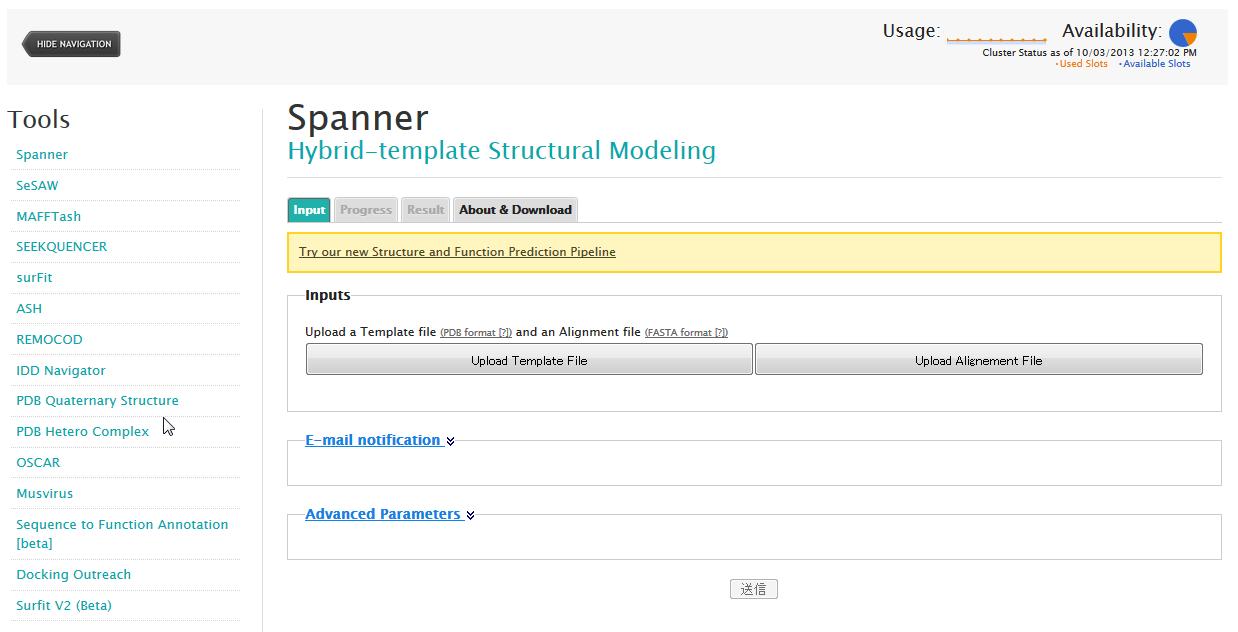 | Spanner Spanner is a structural homology modeling program?that is, it threads a specific amino-acid sequence onto a specific PDB structure, patching up the gaps as best it can. secondary structure, 3D structure, structure prediction |
 | SFAS SFAS (Sequence to Function Annotation Server) is a web-based tool for predicting the structure of an amino acid sequence. SFAS runs several external programs for sequence alignment and structural modeling and organized their results. SFAS does not do any complicated calculations. There are currently several choices for alignment and one choice for structural modeling. In the future, the number of choices will be increased. secondary structure, 3D structure, structure prediction |
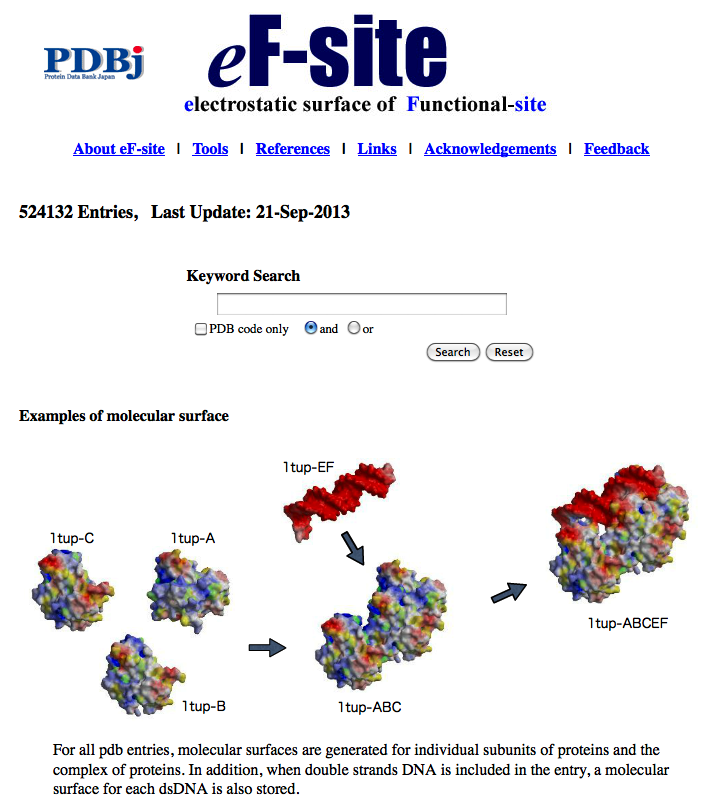 | eF-site eF-site (electrostatic-surface of Functional site) is a database for molecular surfaces of proteins' functional sites, displaying the electrostatic potentials and hydrophobic properties together on the Connolly surfaces of the active sites, for analyses of the molecular recognition mechanisms. 3D structure, function prediction, surface structure |
 | eF-surf eF-surf is a web server to calculate the molecular surface of the up-loaded file with PDB format in the same way as the eF-site database. 3D structure, function prediction, surface structure |
 | eF-seek eF-seek is a web server to search for the similar ligand binding sites for the uploaded coordinate file with PDB format. The representative binding sites in eF-site database are search by our own algorithm based on the clique search algorithm. search, 3D structure, function prediction, surface structure |
 | ProMode Elastic ProMode Elastic is a database of normal mode analysis of PDB data. The normal mode analysis is performed by the program PDBETA we have developed. PDBETA is a program of Elastic-network-model based normal mode analysis in Torsional Angle space for PDB data. PDBETA can describe molecular structures with relatively smaller number of degrees of freedom, and take into computation not only proteins but also DNA, RNA, and ligand molecules (hydrogen atoms and water molecules are excluded currently to suppress the number of variables). 3D structure |
 | eProtS eProtS (Encyclopedia of Protein Structures) is a dictionary with pictures of the protein three-dimensional structures, for understanding the tertiary structures and the biological functions for several selected protein molecules that are particularly important for biology. This Web site can be prepared for the general audience. As it is a wiki-based repository of documents, any people can deposit or edit documents as well as browse. education/dictionary, 3D structure |
 | Molecule of the month Introduction to biological macromolecules written by Dr. David S. Goodsell of RCSB... education/dictionary, 3D structure |
 | Previous workshop Materials used in the previous workshop and tutorials are available from this site. education/dictionary |
 | PDBj Games PDBj Games are a collection of games such as Pelmanism and Snake games through which you can learn about amino acids and proteins, polymers of amino acids. education/dictionary |
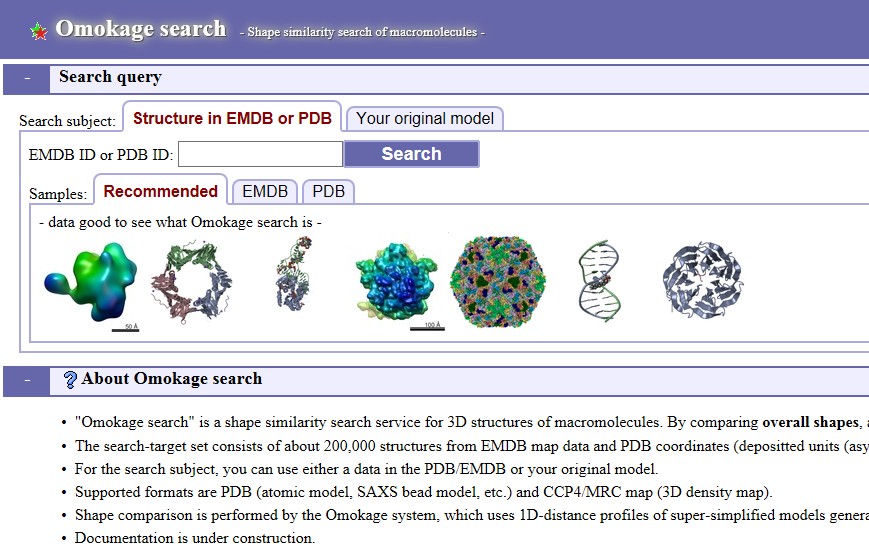 | Omokage search Omokage search is a shape similarity search service for 3D structures of macromolecules. By comparing overall shapes, and ignoring details, similar-shaped structures are searched. PDB, EMDB, electron microscopy, search, 3D structure, similarity |
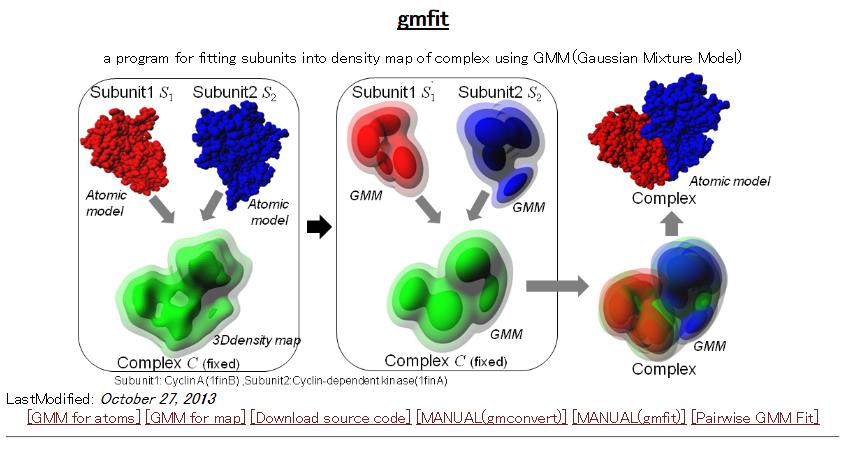 | gmfit gmfit is a program for fast fitting of 3D objects using Gaussian mixture model. The fitting service between two EMDB density maps or PDB structures is available through the WEB. PDB, EMDB, similarity, electron microscopy, 3D structure |
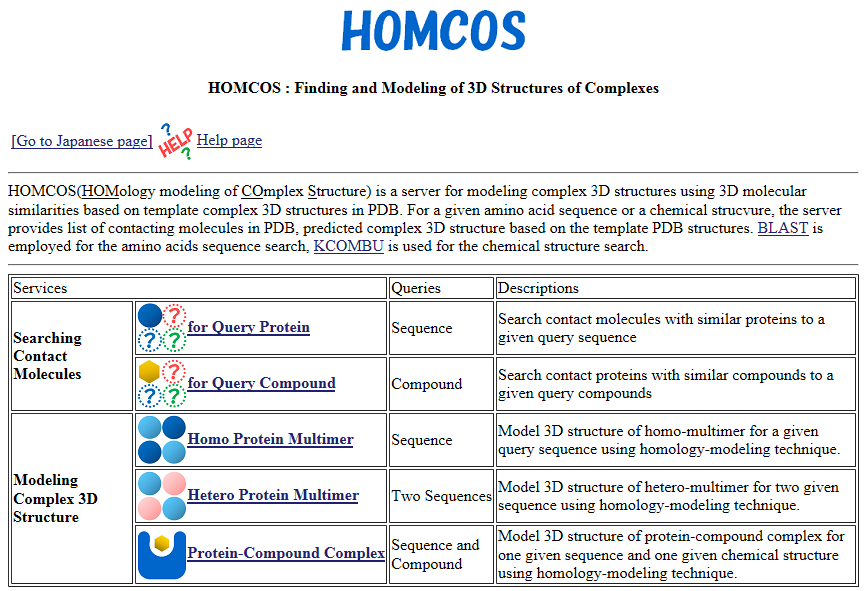 | HOMCOS HOMCOS is a service for searching 3D complex structures in PDB from sequences or chemical structures, and for modeling 3D complex structures based on the found template structures. PDB, search, 3D structure, structure prediction, chemical component, binding site |
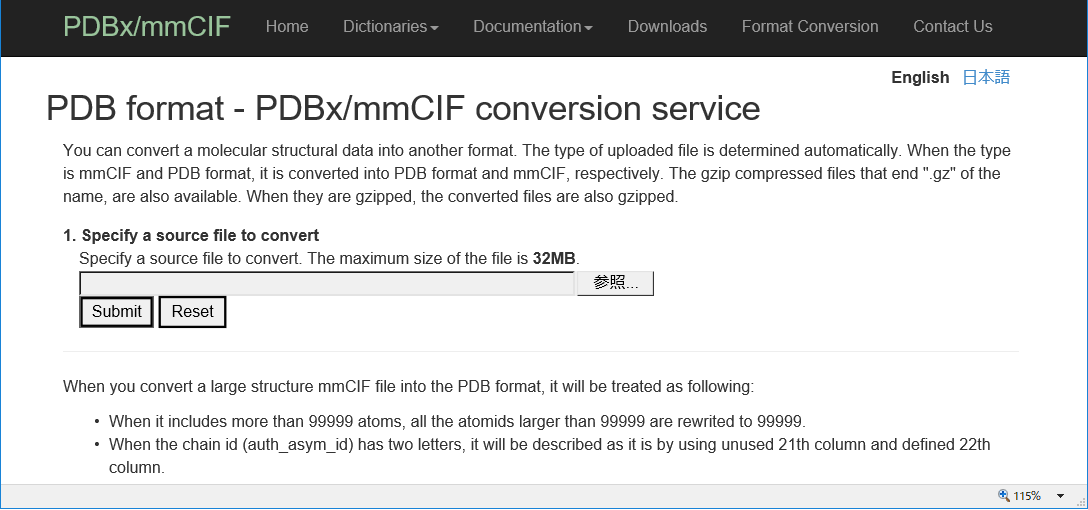 | PDB format - PDBx/mmCIF conversion PDB format - PDBx/mmCIF conversion is a tool for converting into PDB format and mmCIF, respectively. PDB |
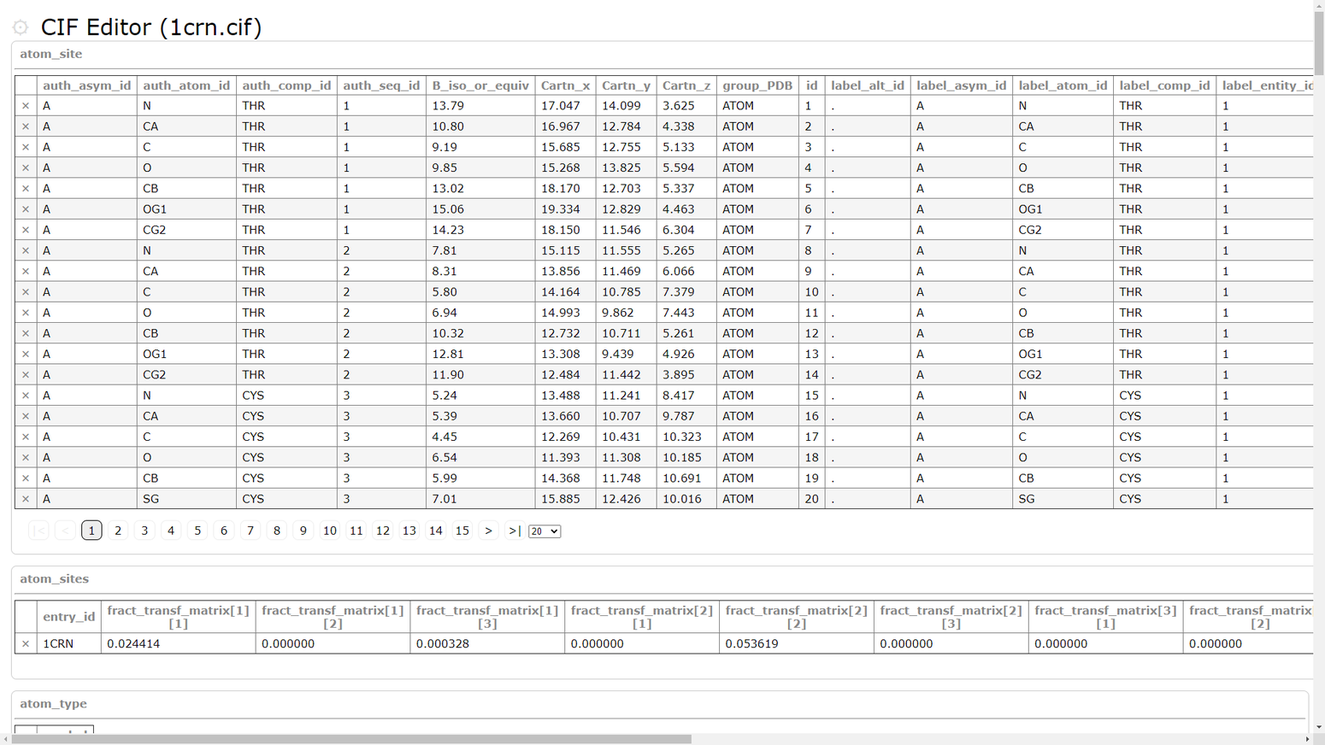 | PDBx/mmCIF editor PDBx/mmCIF editor is a tool for editing PDBx/mmCIF format. PDB |







