6KQV
 
 | | Solution Structure of the UbL Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-08-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
2JAX
 
 | |
7SMI
 
 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea | | Descriptor: | L-galactose dehydrogenase | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-10-26 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
7SML
 
 | | Crystal Structure of L-GALACTONO-1,4-LACTONE DEHYDROGENASE de Myrciaria dubia | | Descriptor: | L-GALACTONO-1,4-LACTONE DEHYDROGENASE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit Camu-Camu.
J.Exp.Bot., 2024
|
|
2DAG
 
 | | Solution Structure of the First UBA Domain in the Human Ubiquitin Specific Protease 5 (Isopeptidase 5) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 5 | | Authors: | Zhao, C, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the First UBA Domain in the Human Ubiquitin Specific Protease 5 (Isopeptidase 5)
To be Published
|
|
2DAK
 
 | | Solution Structure of the Second UBA Domain in the Human Ubiquitin Specific Protease 5 (Isopeptidase 5) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 5 | | Authors: | Zhao, C, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Second UBA Domain in the Human Ubiquitin Specific Protease 5 (Isopeptidase 5)
To be Published
|
|
7SVQ
 
 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea in complex with NAD+ | | Descriptor: | L-galactose dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-11-19 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
7CJM
 
 | | SARS CoV-2 PLpro in complex with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Fu, Z, Huang, H. | | Deposit date: | 2020-07-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
2MUX
 
 | |
3ZLZ
 
 | |
4A3O
 
 | | Crystal structure of the USP15 DUSP-UBL monomer | | Descriptor: | GLYCEROL, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
4A3P
 
 | | Structure of USP15 DUSP-UBL deletion mutant | | Descriptor: | ACETATE ION, IODIDE ION, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
5V5T
 
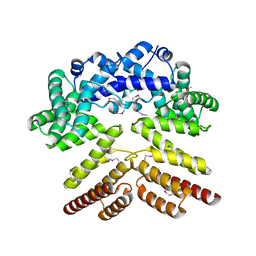 | | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Conserved domain protein | | Authors: | De Groote, M.C.R, Camargo, I.L, Serror, P, Horjales, E. | | Deposit date: | 2017-03-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and functional characterization of ElrR, a member of the RRNPP family of transcriptional regulators.
To be published
|
|
5V5U
 
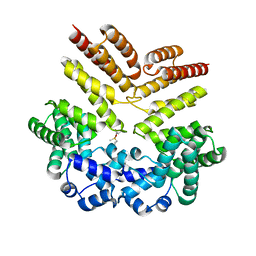 | | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis | | Descriptor: | Conserved domain protein | | Authors: | De Groote, M.C.R, Camargo, I.L, Serror, P, Horjales, E. | | Deposit date: | 2017-03-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis
To be published
|
|
5WFI
 
 | |
3IXP
 
 | |
5OK6
 
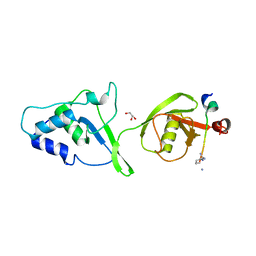 | | Ubiquitin specific protease 11 USP11 - peptide F complex | | Descriptor: | 1,2-ETHANEDIOL, ALA-GLU-GLY-GLU-PHE-TYR-LYS-LEU-LYS-ILE-ARG-THR-PRO-AAR, GLYCEROL, ... | | Authors: | Spiliotopoulos, A, Dreveny, I. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of peptide ligands targeting a specific ubiquitin-like domain-binding site in the deubiquitinase USP11.
J.Biol.Chem., 294, 2019
|
|
3LMN
 
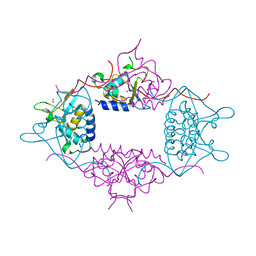 | | Oligomeric structure of the DUSP domain of human USP15 | | Descriptor: | ACETIC ACID, FORMIC ACID, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Walker, J.R, Asinas, A, Avvakumov, G.V, Alenkin, D, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Ubiquitin-Specific Protease 15 DUSP Domain
To be Published
|
|
3OLQ
 
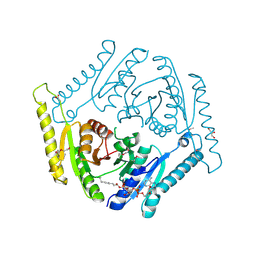 | | The crystal structure of a universal stress protein E from Proteus mirabilis HI4320 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | The crystal structure of a universal stress protein E from
Proteus mirabilis HI4320
To be Published
|
|
3PPA
 
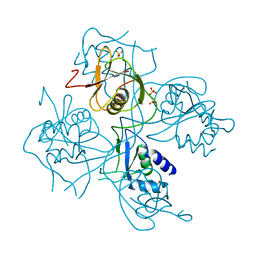 | | Structure of the Dusp-Ubl domains of Usp15 | | Descriptor: | CITRIC ACID, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Walker, J.R, Asinas, A.E, Dong, A, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Dusp-Ubl Domains of the Ubiquitin-Specific Protease 15
To be Published
|
|
3PV1
 
 | | Crystal structure of the USP15 DUSP-UBL domains | | Descriptor: | ACETATE ION, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Elliott, P.R, Urbe, S, Clague, M.J, Barsukov, I.L. | | Deposit date: | 2010-12-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of the ubiquitin specific protease DUSP-UBL double domains.
Febs Lett., 585, 2011
|
|
8DH6
 
 | | Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cytochrome c oxidase subunit 1, ... | | Authors: | Godoy, A.S, Song, Y, Cheruvara, H, Quigley, A, Oliva, G. | | Deposit date: | 2022-06-25 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs
To Be Published
|
|
8DH7
 
 | |
8DKT
 
 | | Crystal Structure of Septin1 - Septin2 heterocomplex from Drosophila melanogaster | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | de Freitas, A.F, Leonardo, D.A, Cavini, I.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2022-07-06 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conservation and divergence of the G-interfaces of Drosophila melanogaster septins.
Cytoskeleton (Hoboken), 80, 2023
|
|
8E25
 
 | | Crystal Structure of SARS-CoV-2 Main Protease M49I mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-08-14 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
