138D
 
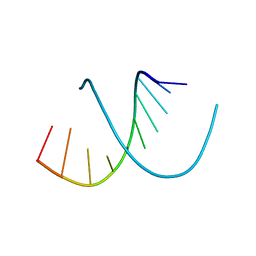 | | A-DNA DECAMER D(GCGGGCCCGC)-HEXAGONAL CRYSTAL FORM | | 分子名称: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | 著者 | Ramakrishnan, B, Sundaralingam, M. | | 登録日 | 1993-09-15 | | 公開日 | 1994-01-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
137D
 
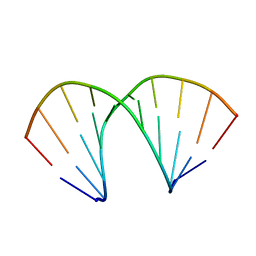 | | A-DNA DECAMER D(GCGGGCCCGC)-ORTHORHOMBIC CRYSTAL FORM | | 分子名称: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | 著者 | Ramakrishnan, B, Sundaralingam, M. | | 登録日 | 1993-09-15 | | 公開日 | 1994-01-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
1LQG
 
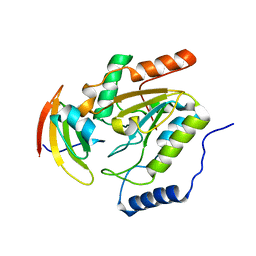 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | 分子名称: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | 登録日 | 2002-05-10 | | 公開日 | 2002-11-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LQJ
 
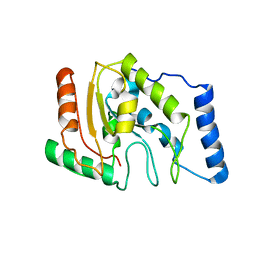 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | 分子名称: | URACIL-DNA GLYCOSYLASE | | 著者 | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | 登録日 | 2002-05-10 | | 公開日 | 2002-11-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4EE5
 
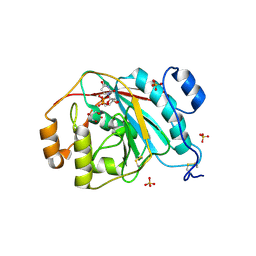 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with trisaccharide from Lacto-N-neotetraose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | 著者 | Ramakrishnan, B, Qasba, P.K. | | 登録日 | 2012-03-28 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
4EEM
 
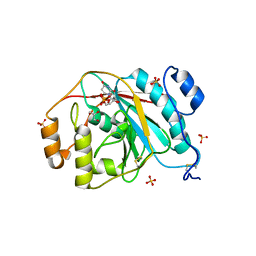 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-MAN-ALPHA-methyl | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-methyl alpha-D-mannopyranoside, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | 著者 | Ramakrishnan, B, Qasba, P.K. | | 登録日 | 2012-03-28 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
4EE3
 
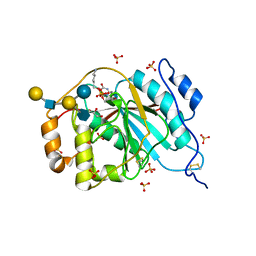 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with pentasaccharide | | 分子名称: | 6-AMINOHEXYL-URIDINE-C1,5'-DIPHOSPHATE, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | 著者 | Ramakrishnan, B, Qasba, P.K. | | 登録日 | 2012-03-28 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
5CBG
 
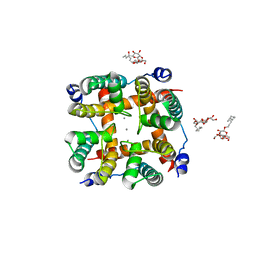 | | Calcium activated non-selective cation channel | | 分子名称: | CALCIUM ION, DECYL-BETA-D-MALTOPYRANOSIDE, Ion transport 2 domain protein | | 著者 | Dhakshnamoorthy, B, Rohaim, A, Rui, H, Blachowicz, L, Roux, B. | | 登録日 | 2015-06-30 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Structural and functional characterization of a calcium-activated cation channel from Tsukamurella paurometabola.
Nat Commun, 7, 2016
|
|
5C9S
 
 | |
7O3O
 
 | | Structure of haloalkane dehalogenase mutant DhaA80(T148L, G171Q, A172V, C176F) from Rhodococcus rhodochrous with ionic liquid | | 分子名称: | CHLORIDE ION, ETHANOLAMINE, Haloalkane dehalogenase | | 著者 | Shaposhnikova, A, Prudnikova, T, Kuta Smatanova, I. | | 登録日 | 2021-04-02 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Stabilization of Haloalkane Dehalogenase Structure by Interfacial Interaction with Ionic Liquids
Crystals, 11, 2021
|
|
7O8B
 
 | |
5Z19
 
 | |
5Z1A
 
 | |
5Z18
 
 | |
5Z1B
 
 | |
1WQG
 
 | | Crystal structure of ribosome recycling factor from Mycobacterium Tuberculosis | | 分子名称: | CADMIUM ION, Ribosome recycling factor | | 著者 | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | 登録日 | 2004-09-29 | | 公開日 | 2005-02-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
1WQF
 
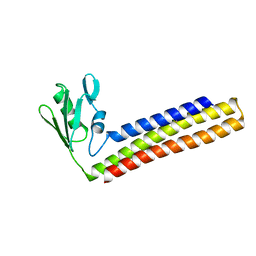 | | Crystal structure of Ribosome recycling factor from Mycobacterium Tuberculosis | | 分子名称: | CADMIUM ION, Ribosome recycling factor | | 著者 | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | 登録日 | 2004-09-28 | | 公開日 | 2005-02-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
1WQH
 
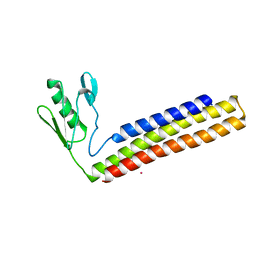 | | Crystal structure of ribosome recycling factor from Mycobacterium tuberculosis | | 分子名称: | CADMIUM ION, Ribosome recycling factor | | 著者 | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | 登録日 | 2004-09-29 | | 公開日 | 2005-02-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | X-ray structural studies of Mycobacterium Tuberculosis RRF and a comparative study of RRFS of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
1X3E
 
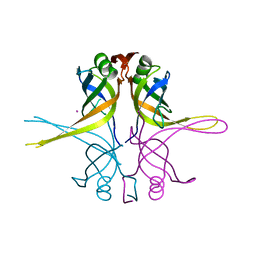 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium smegmatis | | 分子名称: | CADMIUM ION, Single-strand binding protein | | 著者 | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | 登録日 | 2005-05-04 | | 公開日 | 2005-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X3G
 
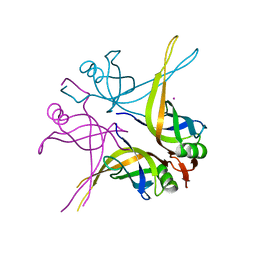 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | 分子名称: | CADMIUM ION, Single-strand binding protein | | 著者 | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | 登録日 | 2005-05-05 | | 公開日 | 2005-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X3F
 
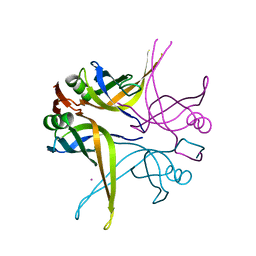 | | Crystal structure of the single-stranded DNA-binding protein from Mycobacterium SMEGMATIS | | 分子名称: | CADMIUM ION, Single-strand binding protein | | 著者 | Saikrishnan, K, Manjunath, G.P, Singh, P, Jeyakanthan, J, Dauter, Z, Sekar, K, Muniyappa, K, Vijayan, M. | | 登録日 | 2005-05-05 | | 公開日 | 2005-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7VSR
 
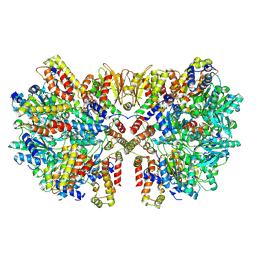 | | Structure of McrBC (stalkless mutant) | | 分子名称: | 5-methylcytosine-specific restriction enzyme B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Saikrishnan, K, Adhav, V.A, Bose, S, Kutti R, V. | | 登録日 | 2021-10-27 | | 公開日 | 2023-02-15 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure of McrBC (stalkless mutant)
To Be Published
|
|
3U44
 
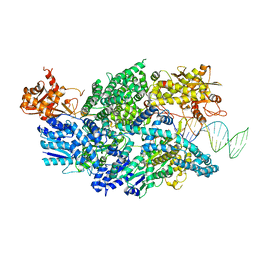 | | Crystal structure of AddAB-DNA complex | | 分子名称: | ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, DNA (36-MER), ... | | 著者 | Saikrishnan, K, Krajewski, W, Wigley, D. | | 登録日 | 2011-10-07 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.201 Å) | | 主引用文献 | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
149D
 
 | |
4RND
 
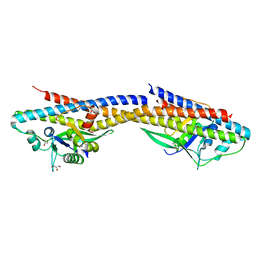 | |
