1I1W
 
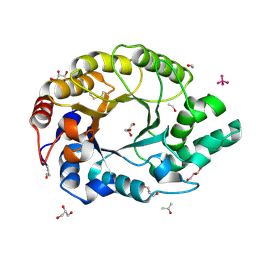 | | 0.89A Ultra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus | | Descriptor: | ACETONE, ENDO-1,4-BETA-XYLANASE, ETHANOL, ... | | Authors: | Natesh, R, Ramakumar, S, Viswamitra, M.A. | | Deposit date: | 2001-02-04 | | Release date: | 2003-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Thermostable xylanase from Thermoascus aurantiacus at ultrahigh resolution (0.89 A) at 100 K and atomic resolution (1.11 A) at 293 K refined anisotropically to small-molecule accuracy.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
8R5K
 
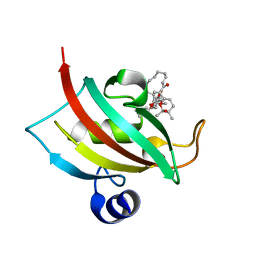 | |
1OB7
 
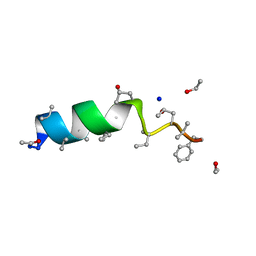 | | Cephaibol C | | Descriptor: | CEPHAIBOL C, ETHANOL, SODIUM ION | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
1OB6
 
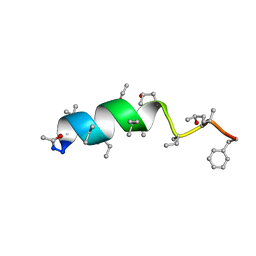 | | Cephaibol B | | Descriptor: | ACETATE ION, CEPHAIBOL B, ETHANOL | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
1ETN
 
 | |
1ENN
 
 | | SOLVENT ORGANIZATION IN AN OLIGONUCLEOTIDE CRYSTAL: THE STRUCTURE OF D(GCGAATTCG)2 AT ATOMIC RESOLUTION | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Soler-Lopez, M, Malinina, L, Subirana, J.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Solvent organization in an oligonucleotide crystal. The structure of d(GCGAATTCG)2 at atomic resolution.
J.Biol.Chem., 275, 2000
|
|
4UA7
 
 | |
1JXW
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 180 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4U9H
 
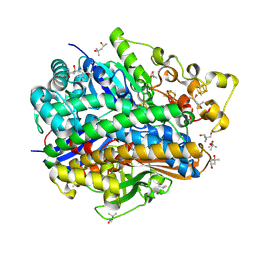 | | Ultra High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Nishikawa, K, Lubitz, W. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
1JXT
 
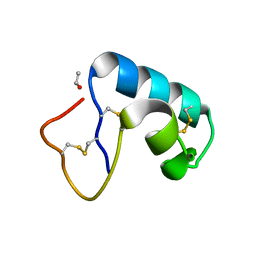 | | CRAMBIN MIXED SEQUENCE FORM AT 160 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3IP0
 
 | |
1JXX
 
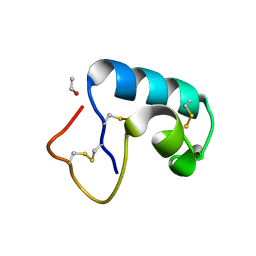 | | CRAMBIN MIXED SEQUENCE FORM AT 200 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXY
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 220 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3UI6
 
 | | 0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SODIUM ION, ... | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | 0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT
To be Published
|
|
4WEE
 
 | |
6KM2
 
 | | Human Carbonic Anhydrase II V143I variant 15 atm CO2 | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | Kim, C.U, Kim, J.K. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Iucrj, 7, 2020
|
|
7TWW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 6 (P43 crystal form) | | Descriptor: | CITRIC ACID, Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-02-07 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 6 (P43 crystal form)
To Be Published
|
|
1ET1
 
 | | CRYSTAL STRUCTURE OF HUMAN PARATHYROID HORMONE 1-34 AT 0.9 A RESOLUTION | | Descriptor: | PARATHYROID HORMONE, SODIUM ION | | Authors: | Jin, L, Briggs, S.L, Chandrasekhar, S, Chirgadze, N.Y, Clawson, D.K, Schevitz, R.W, Smiley, D.L, Tashjian, A.H, Zhang, F. | | Deposit date: | 2000-04-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of human parathyroid hormone 1-34 at 0.9-A resolution.
J.Biol.Chem., 275, 2000
|
|
7TWJ
 
 | |
7TWN
 
 | |
7TWS
 
 | |
7TWX
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 7 (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-02-07 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 7 (P43 crystal form)
To Be Published
|
|
7TWR
 
 | |
7TWQ
 
 | |
7TWT
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 4 (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-02-07 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 4 (P43 crystal form)
To Be Published
|
|
