7TVM
 
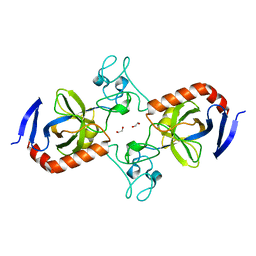 | | Viral AMG chitosanase V-Csn, apo structure, crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7VPA
 
 | | Crystal structure of Ple629 from marine microbial consortium | | Descriptor: | hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | Descriptor: | CALCIUM ION, hydrolase Ple628 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7RKA
 
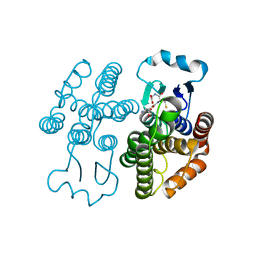 | |
7TOG
 
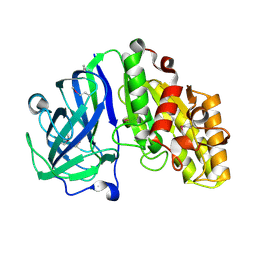 | | Crystal structure of carbohydrate esterase PbeAcXE, apoenzyme | | Descriptor: | SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOK
 
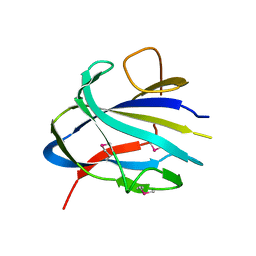 | | Crystal structure of the CBM domain of carbohydrate esterase FjoAcXE | | Descriptor: | Acetylxylan esterase I | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOJ
 
 | | Crystal structure of carbohydrate esterase CspAcXE, apoenzyme | | Descriptor: | CHLORIDE ION, SGNH/GDSL hydrolase family protein | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOH
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with MeGlcpA-Xylp | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose, SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOI
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with acetate | | Descriptor: | ACETATE ION, SGNH hydrolase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7KTR
 
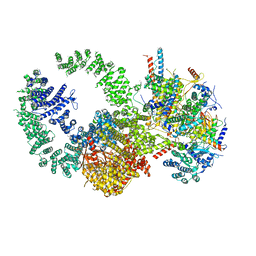 | | Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) | | Descriptor: | Ataxin-7, INOSITOL HEXAKISPHOSPHATE, Isoform 3 of Transcription factor SPT20 homolog, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KTS
 
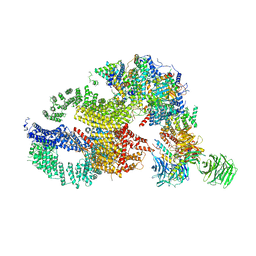 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | Descriptor: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (19.09 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7A3W
 
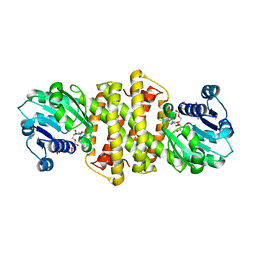 | | Structure of Imine Reductase from Pseudomonas sp. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NAD(P)-dependent oxidoreductase, ... | | Authors: | Cuetos, A, Thorpe, T, Turner, N.J, Grogan, G. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Multifunctional biocatalyst for conjugate reduction and reductive amination.
Nature, 604, 2022
|
|
7JJV
 
 | |
6WQ0
 
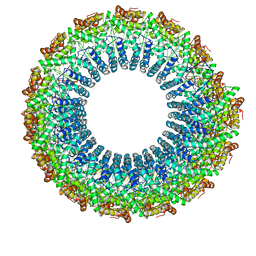 | | Cryo-EM of the S. solfataricus rod-shaped virus, SSRV1 | | Descriptor: | DNA (301-MER), Structural protein | | Authors: | Wang, F, Baquero, D.P, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5ZFG
 
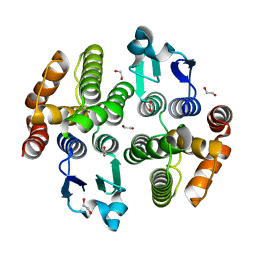 | | Crystal structure of a diazinon-metabolizing glutathione S-transferase in the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterisation of a diazinon-metabolising glutathione S-transferase in the silkworm Bombyx mori by X-ray crystallography and genome editing analysis.
Sci Rep, 8, 2018
|
|
5KGF
 
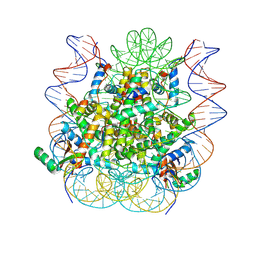 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
5FJQ
 
 | | Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus | | Descriptor: | CARBOHYDRATE BINDING PROTEIN, PUTATIVE, CPB33A, ... | | Authors: | Forsberg, Z, Nelson, C.E, Dalhus, B, Mekasha, S, Loose, J.S.M, Rohr, A.K, Eijsink, V.G.H, Gardner, J.G, Vaaje-Kolstad, G. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Analysis of a Lytic Polysaccharide Monooxygenase Important for Efficient Utilization of Chitin in Cellvibrio Japonicus
J.Biol.Chem., 291, 2016
|
|
5CBX
 
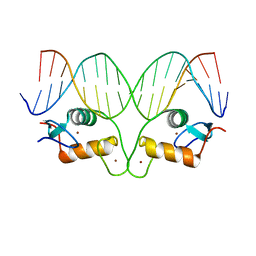 | | AncGR DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CBY
 
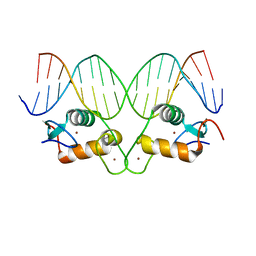 | | AncGR2 DNA Binding Domain - (+)GRE Complex | | Descriptor: | AncGR2 DNA Binding Domain, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2MJM
 
 | |
4MB8
 
 | |
4J95
 
 | |
3AY8
 
 | | Glutathione S-transferase unclassified 2 from Bombyx mori | | Descriptor: | GLYCEROL, Glutathione S-transferase | | Authors: | Kakuta, Y, Usuda, K, Nakashima, T, Kimura, M, Aso, Y, Yamamoto, K. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic survey of active sites of an unclassified glutathione transferase from Bombyx mori
Biochim.Biophys.Acta, 1810, 2011
|
|
