5N5C
 
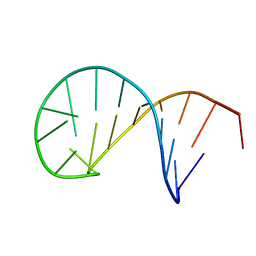 | | NMR solution structure of the TSL2 RNA hairpin | | Descriptor: | RNA (19-MER) | | Authors: | Garcia-Lopez, A, Wacker, A, Tessaro, F, Jonker, H.R.A, Richter, C, Comte, A, Berntenis, N, Schmucki, R, Hatje, K, Sciarra, D, Konieczny, P, Fournet, G, Faustino, I, Orozco, M, Artero, R, Goekjian, P, Metzger, F, Ebeling, M, Joseph, B, Schwalbe, H, Scapozza, L. | | Deposit date: | 2017-02-13 | | Release date: | 2018-03-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes.
Nat Commun, 9, 2018
|
|
7D58
 
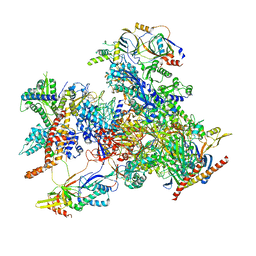 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DN3
 
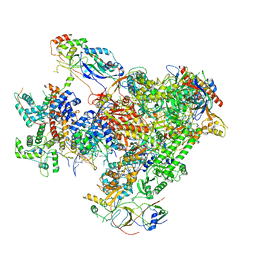 | | Structure of Human RNA Polymerase III elongation complex | | Descriptor: | DNA (5'-D(P*TP*CP*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*TP*CP*GP*GP*G)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
1AL5
 
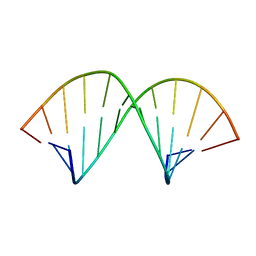 | | A-TRACT RNA DODECAMER, NMR, 12 STRUCTURES | | Descriptor: | RNA (5'-R(*CP*GP*CP*AP*AP*AP*UP*UP*UP*GP*CP*G)-3') | | Authors: | Conte, M.R, Conn, G.L, Brown, T, Lane, A.N. | | Deposit date: | 1997-06-11 | | Release date: | 1997-12-17 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Conformational properties and thermodynamics of the RNA duplex r(CGCAAAUUUGCG)2: comparison with the DNA analogue d(CGCAAATTTGCG)2.
Nucleic Acids Res., 25, 1997
|
|
6UPZ
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UPY
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 2E | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UQ3
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 5 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UPX
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UQ2
 
 | | RNA polymerase II elongation complex with dG in state 1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UQ0
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 4 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UQ1
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 6 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1I3Q
 
 | | RNA POLYMERASE II CRYSTAL FORM I AT 3.1 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-15 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1I50
 
 | | RNA POLYMERASE II CRYSTAL FORM II AT 2.8 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-23 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
7KEF
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaM in swing state | | Descriptor: | (1S)-1,4-anhydro-1-(3-methoxynaphthalen-2-yl)-5-O-phosphono-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KED
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, W, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KEE
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaMTP bound to E-site | | Descriptor: | (1S)-1,4-anhydro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-1-(3-methoxynaphthalen-2-yl)-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
5G5L
 
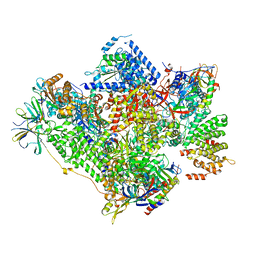 | | RNA polymerase I-Rrn3 complex at 4.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-27 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | RNA Polymerase I-Rrn3 Complex at 4.8 A Resolution
Nat.Commun., 7, 2016
|
|
1BVJ
 
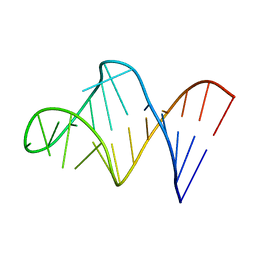 | | HIV-1 RNA A-RICH HAIRPIN LOOP | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*AP*CP*GP*GP*UP*GP*UP*AP*AP*AP*AP*AP*UP*CP*UP*CP*GP*CP* C)-3') | | Authors: | Puglisi, E.V, Puglisi, J.D. | | Deposit date: | 1998-08-31 | | Release date: | 1998-12-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | HIV-1 A-rich RNA loop mimics the tRNA anticodon structure.
Nat.Struct.Biol., 5, 1998
|
|
6GBM
 
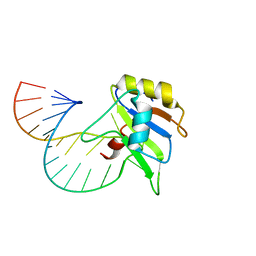 | | Solution structure of FUS-RRM bound to stem-loop RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*GP*AP*UP*UP*AP*CP*AP*AP*UP*UP*CP*UP*AP*UP*UP*UP*GP*CP*C)-3'), RNA-binding protein FUS | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
7OB9
 
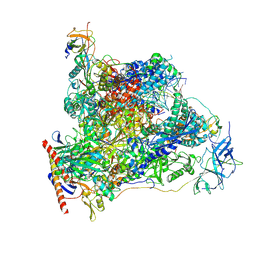 | | Cryo-EM structure of human RNA Polymerase I in elongation state | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
157D
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF R(CGCGAAUUAGCG): AN RNA DUPLEX CONTAINING TWO G(ANTI).A(ANTI) BASE-PAIRS | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*AP*GP*CP*G)-3') | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Ebel, S, Lough, D.M, Brown, T, Hunter, W.N. | | Deposit date: | 1994-02-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and molecular structure of r(CGCGAAUUAGCG): an RNA duplex containing two G(anti).A(anti) base pairs.
Structure, 2, 1994
|
|
5LSN
 
 | |
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
